Creates lollipop charts for categorical data visualization following R Graph Gallery best practices, with emphasis on clinical applications like patient timelines, treatment outcomes, and biomarker comparisons. Uses geom_segment() and geom_point() for optimal visual presentation.
Usage
lollipop(
data,
dep,
group,
useHighlight = FALSE,
highlight = "",
aggregation = "none",
sortBy = "original",
orientation = "vertical",
showValues = FALSE,
showMean = FALSE,
colorScheme = "default",
theme = "default",
pointSize = 3,
lineWidth = 1,
lineType = "solid",
baseline = 0,
conditionalColor = FALSE,
colorThreshold = 0,
xlabel = "",
ylabel = "",
title = "",
width = 800,
height = 600
)Arguments
- data
The data as a data frame.
- dep
The numeric variable for the values (lollipop heights/lengths).
- group
The categorical variable for grouping (lollipop categories).
- useHighlight
Enable or disable highlighting of specific levels in the plot.
- highlight
Specific level to highlight in the plot with different color/style. Only used when useHighlight is TRUE.
- aggregation
How to aggregate multiple values per group. Use 'Mean' or 'Median' for typical clinical measurements, 'Sum' for counts. 'No Aggregation' will over-plot if multiple rows per group exist.
- sortBy
How to sort the lollipops in the chart.
- orientation
Chart orientation (vertical or horizontal lollipops).
- showValues
Whether to display value labels on the lollipops.
- showMean
Whether to display a reference line at the mean value.
- colorScheme
Color scheme for the lollipops.
- theme
Overall theme/appearance of the plot.
- pointSize
Size of the lollipop points.
- lineWidth
Width of the lollipop stems.
- lineType
Type of line for lollipop stems.
- baseline
Starting point for lollipop stems (default is 0).
- conditionalColor
Enable coloring based on value thresholds.
- colorThreshold
Threshold value for conditional coloring (values above/below get different colors).
- xlabel
Custom label for the x-axis.
- ylabel
Custom label for the y-axis.
- title
Custom title for the plot.
- width
Width of the plot in pixels.
- height
Height of the plot in pixels.
Value
A results object containing:
results$todo | a html | ||||
results$summary | a table | ||||
results$plot | an image |
Tables can be converted to data frames with asDF or as.data.frame. For example:
results$summary$asDF
as.data.frame(results$summary)
Examples
# Load clinical lab data
clinical_lab_data <- read.csv("clinical_lab_data.csv")
#> Warning: cannot open file 'clinical_lab_data.csv': No such file or directory
#> Error in file(file, "rt"): cannot open the connection
# Or load from package: data("clinical_lab_data", package = "ClinicoPath")
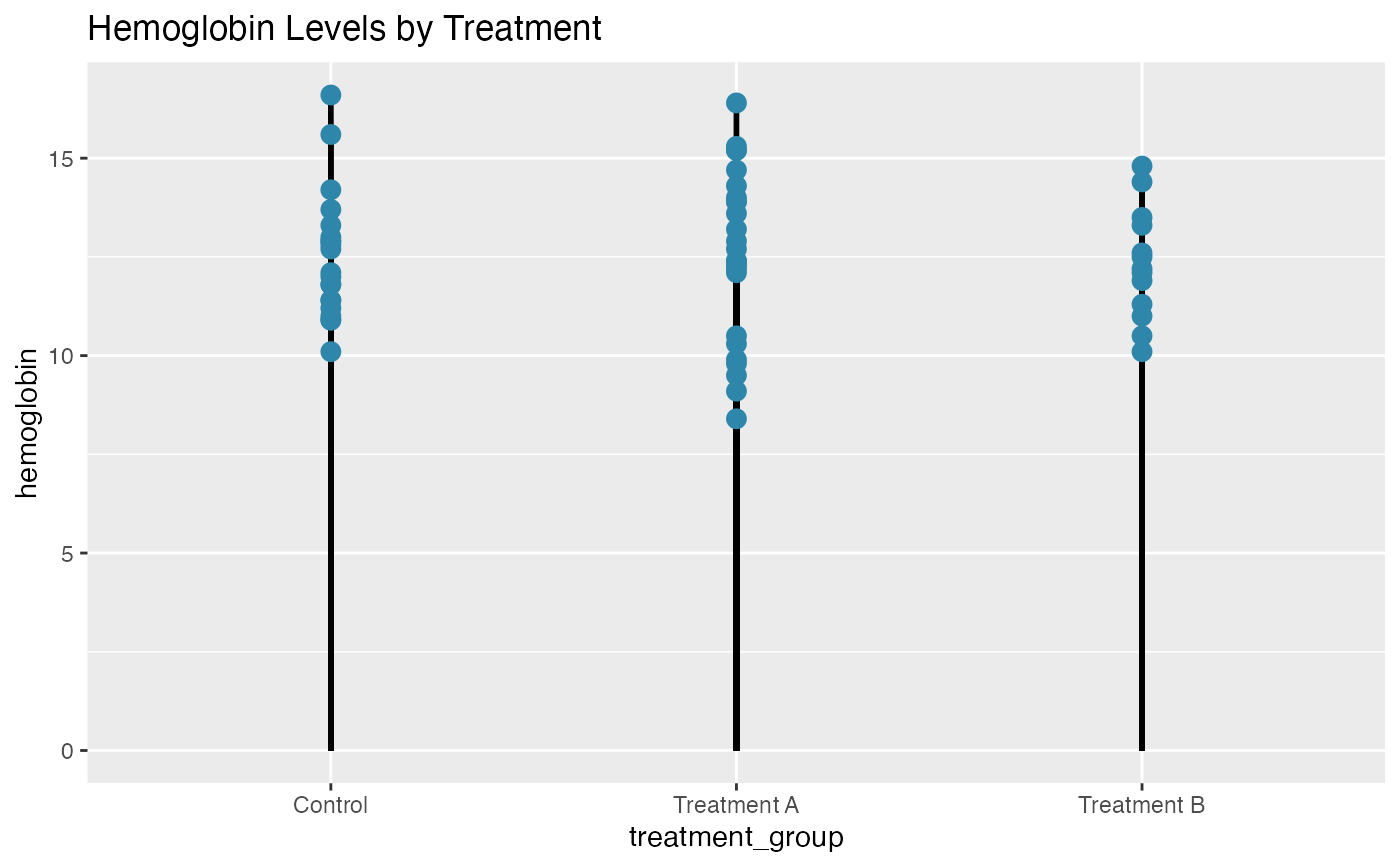
# Basic lollipop chart - Hemoglobin by treatment group
lollipop(
data = clinical_lab_data,
dep = "hemoglobin",
group = "treatment_group",
sortBy = "value_desc",
title = "Hemoglobin Levels by Treatment"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=26 per group). Groups
#> with duplicates: Control, Treatment A, Treatment B. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 12.48 (Standard Deviation = 1.79)Value range: 8.4 -
#> 16.6Highest values found in: Treatment ALowest values found in:
#> Treatment B
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ─────────────────────────────────────────
#> Statistic Value
#> ─────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 12.483333
#> Median Value 12.400000
#> Standard Deviation 1.793129
#> Value Range 8.4 - 16.6
#> Highest Value Group Treatment A
#> Lowest Value Group Treatment B
#> ─────────────────────────────────────────
#>
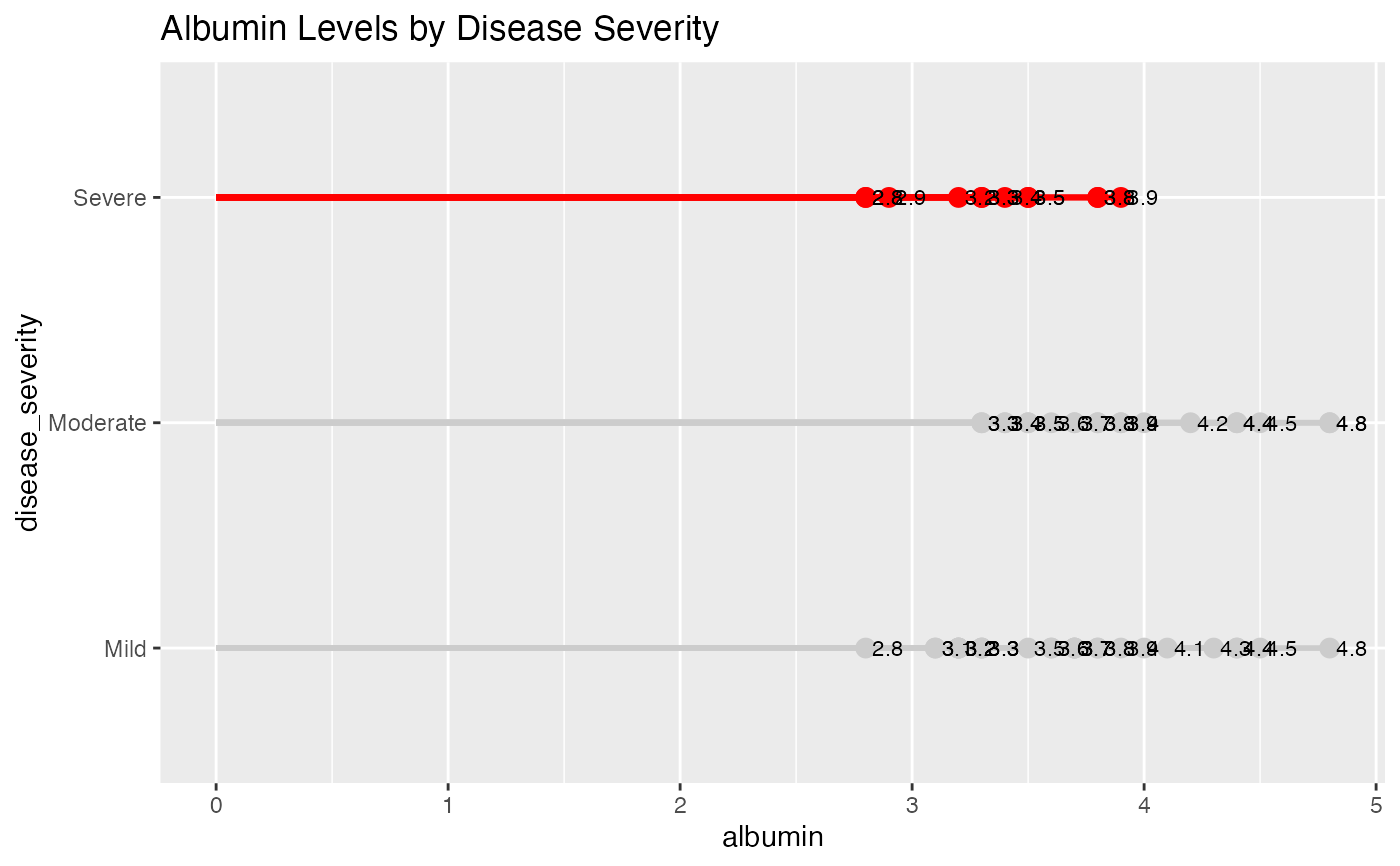
 # Highlighting severe disease cases - Albumin levels
lollipop(
data = clinical_lab_data,
dep = "albumin",
group = "disease_severity",
useHighlight = TRUE,
highlight = "Severe",
orientation = "horizontal",
showValues = TRUE,
colorScheme = "clinical",
title = "Albumin Levels by Disease Severity"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=28 per group). Groups
#> with duplicates: Mild, Moderate, Severe. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 3.67 (Standard Deviation = 0.5)Value range: 2.8 -
#> 4.8Highest values found in: ModerateLowest values found in: Severe
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ───────────────────────────────────────
#> Statistic Value
#> ───────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 3.6750000
#> Median Value 3.6500000
#> Standard Deviation 0.5027467
#> Value Range 2.8 - 4.8
#> Highest Value Group Moderate
#> Lowest Value Group Severe
#> ───────────────────────────────────────
#>
# Highlighting severe disease cases - Albumin levels
lollipop(
data = clinical_lab_data,
dep = "albumin",
group = "disease_severity",
useHighlight = TRUE,
highlight = "Severe",
orientation = "horizontal",
showValues = TRUE,
colorScheme = "clinical",
title = "Albumin Levels by Disease Severity"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=28 per group). Groups
#> with duplicates: Mild, Moderate, Severe. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 3.67 (Standard Deviation = 0.5)Value range: 2.8 -
#> 4.8Highest values found in: ModerateLowest values found in: Severe
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ───────────────────────────────────────
#> Statistic Value
#> ───────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 3.6750000
#> Median Value 3.6500000
#> Standard Deviation 0.5027467
#> Value Range 2.8 - 4.8
#> Highest Value Group Moderate
#> Lowest Value Group Severe
#> ───────────────────────────────────────
#>
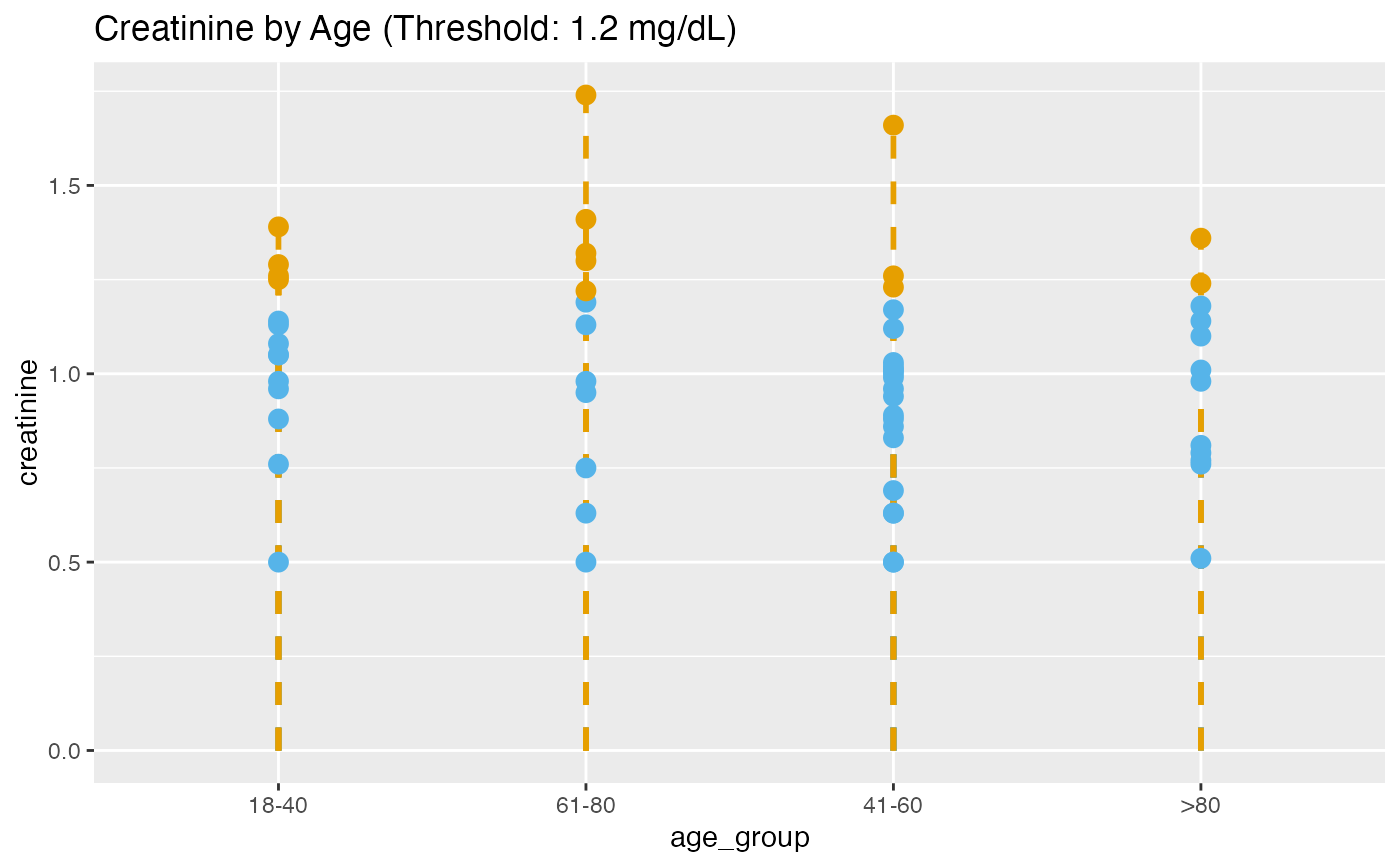
 # Conditional coloring for abnormal creatinine (>1.2 mg/dL)
lollipop(
data = clinical_lab_data,
dep = "creatinine",
group = "age_group",
conditionalColor = TRUE,
colorThreshold = 1.2,
lineType = "dashed",
sortBy = "value_asc",
title = "Creatinine by Age (Threshold: 1.2 mg/dL)"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=22 per group). Groups
#> with duplicates: 18-40, 41-60, 61-80, >80. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> Conditional coloring applied. Values > 1.20 colored orange (above
#> threshold), others blue (below).
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 4
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 1 (Standard Deviation = 0.27)Value range: 0.5 -
#> 1.74Highest values found in: 61-80Lowest values found in: 41-60
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ────────────────────────────────────────
#> Statistic Value
#> ────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 4
#> Mean Value 1.0050000
#> Median Value 1.0100000
#> Standard Deviation 0.2740191
#> Value Range 0.5 - 1.74
#> Highest Value Group 61-80
#> Lowest Value Group 41-60
#> ────────────────────────────────────────
#>
# Conditional coloring for abnormal creatinine (>1.2 mg/dL)
lollipop(
data = clinical_lab_data,
dep = "creatinine",
group = "age_group",
conditionalColor = TRUE,
colorThreshold = 1.2,
lineType = "dashed",
sortBy = "value_asc",
title = "Creatinine by Age (Threshold: 1.2 mg/dL)"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=22 per group). Groups
#> with duplicates: 18-40, 41-60, 61-80, >80. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> Conditional coloring applied. Values > 1.20 colored orange (above
#> threshold), others blue (below).
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 4
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 1 (Standard Deviation = 0.27)Value range: 0.5 -
#> 1.74Highest values found in: 61-80Lowest values found in: 41-60
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ────────────────────────────────────────
#> Statistic Value
#> ────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 4
#> Mean Value 1.0050000
#> Median Value 1.0100000
#> Standard Deviation 0.2740191
#> Value Range 0.5 - 1.74
#> Highest Value Group 61-80
#> Lowest Value Group 41-60
#> ────────────────────────────────────────
#>
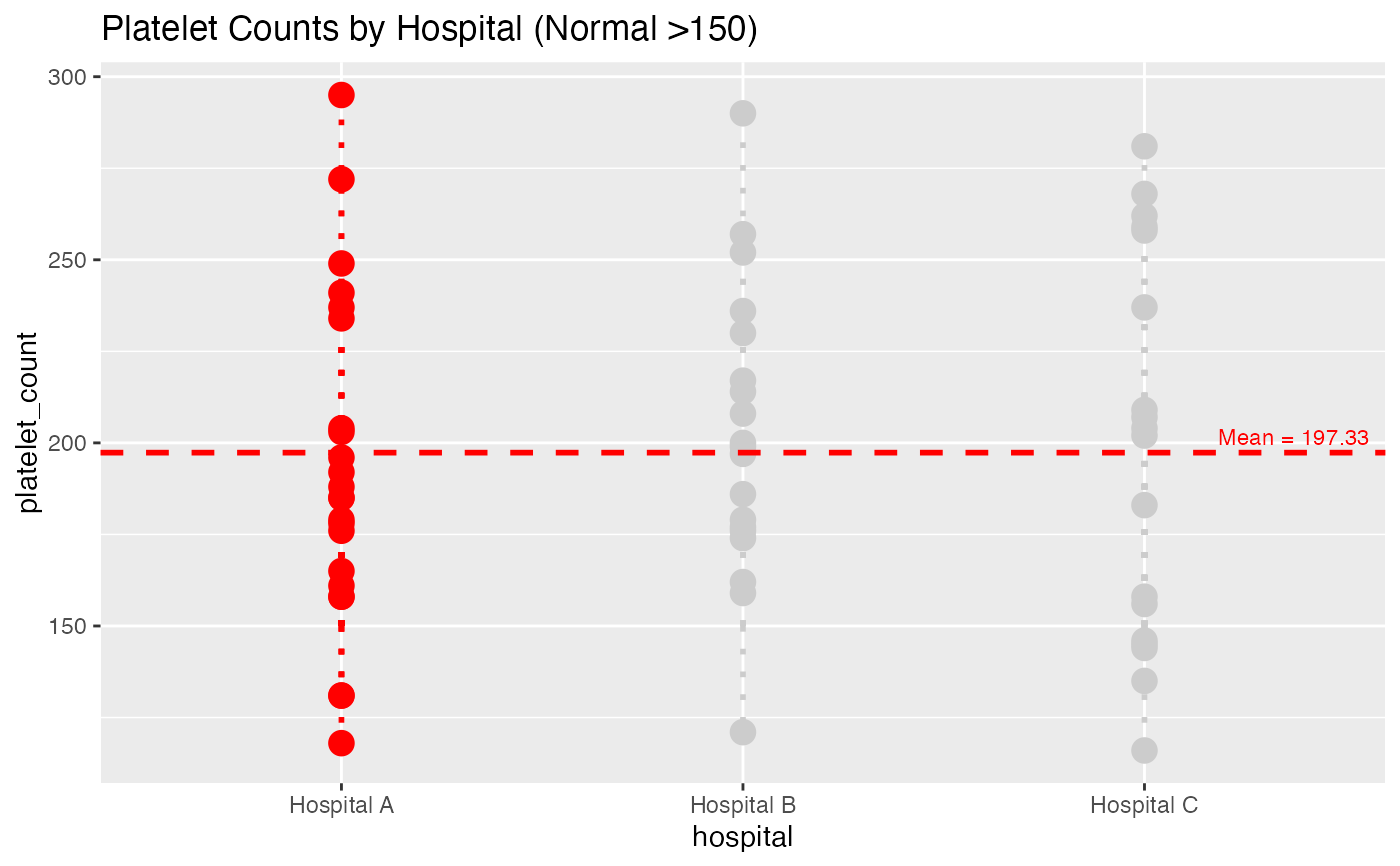
 # Advanced - Platelet count with clinical baseline
lollipop(
data = clinical_lab_data,
dep = "platelet_count",
group = "hospital",
baseline = 150, # Lower normal limit
useHighlight = TRUE,
highlight = "Hospital A",
lineType = "dotted",
pointSize = 4,
showMean = TRUE,
colorScheme = "clinical",
title = "Platelet Counts by Hospital (Normal >150)"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=23 per group). Groups
#> with duplicates: Hospital A, Hospital B, Hospital C. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 197.33 (Standard Deviation = 45.37)Value range: 116 -
#> 295Highest values found in: Hospital BLowest values found in: Hospital
#> A
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ────────────────────────────────────────
#> Statistic Value
#> ────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 197.33333
#> Median Value 194.00000
#> Standard Deviation 45.36880
#> Value Range 116 - 295
#> Highest Value Group Hospital B
#> Lowest Value Group Hospital A
#> ────────────────────────────────────────
#>
# Advanced - Platelet count with clinical baseline
lollipop(
data = clinical_lab_data,
dep = "platelet_count",
group = "hospital",
baseline = 150, # Lower normal limit
useHighlight = TRUE,
highlight = "Hospital A",
lineType = "dotted",
pointSize = 4,
showMean = TRUE,
colorScheme = "clinical",
title = "Platelet Counts by Hospital (Normal >150)"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=23 per group). Groups
#> with duplicates: Hospital A, Hospital B, Hospital C. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 197.33 (Standard Deviation = 45.37)Value range: 116 -
#> 295Highest values found in: Hospital BLowest values found in: Hospital
#> A
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ────────────────────────────────────────
#> Statistic Value
#> ────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 197.33333
#> Median Value 194.00000
#> Standard Deviation 45.36880
#> Value Range 116 - 295
#> Highest Value Group Hospital B
#> Lowest Value Group Hospital A
#> ────────────────────────────────────────
#>
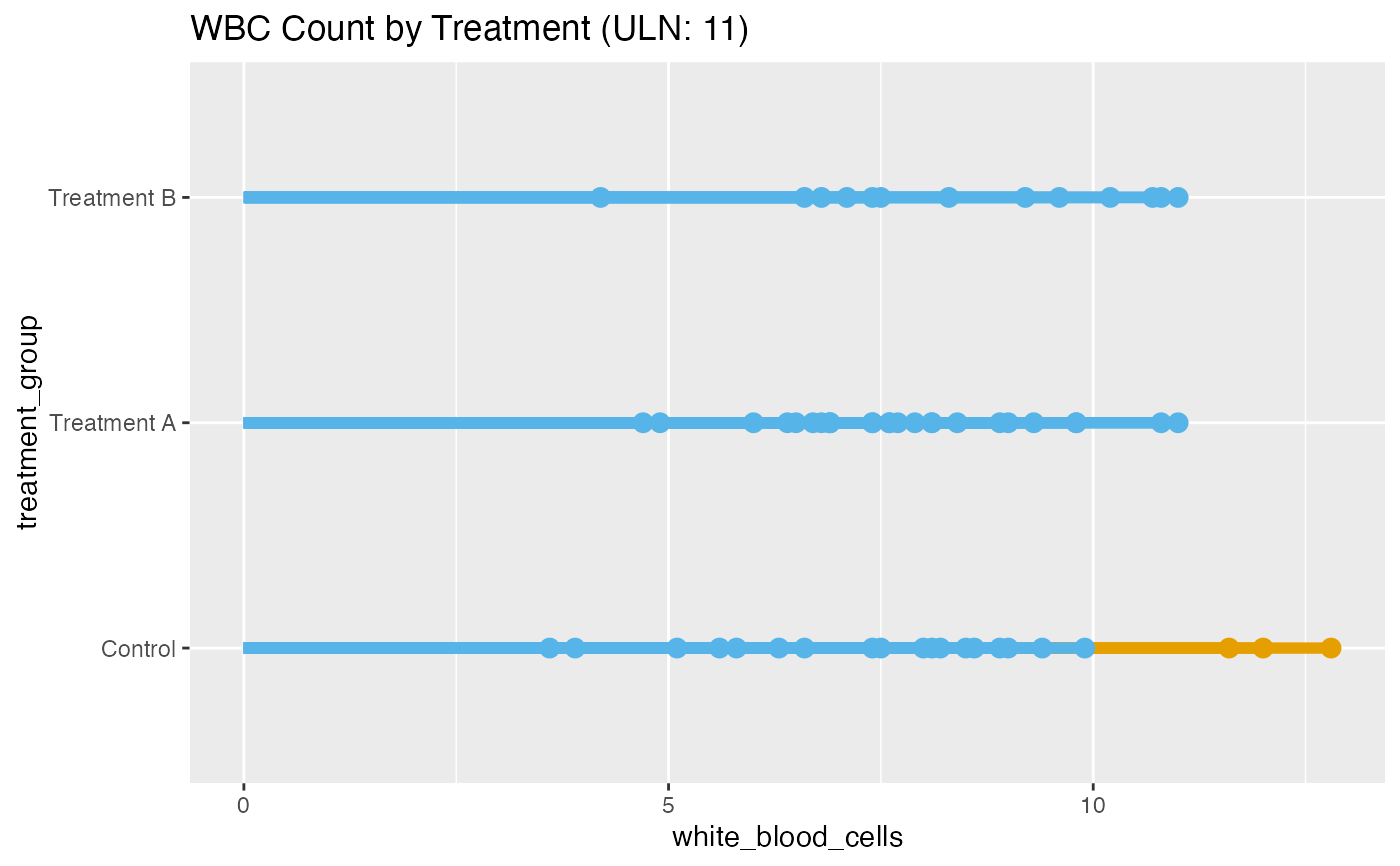
 # Compare WBC across multiple factors
lollipop(
data = clinical_lab_data,
dep = "white_blood_cells",
group = "treatment_group",
conditionalColor = TRUE,
colorThreshold = 11, # Upper normal limit
orientation = "horizontal",
sortBy = "value_desc",
lineWidth = 2,
title = "WBC Count by Treatment (ULN: 11)"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=26 per group). Groups
#> with duplicates: Control, Treatment A, Treatment B. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> Conditional coloring applied. Values > 11.00 colored orange (above
#> threshold), others blue (below).
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 7.96 (Standard Deviation = 2)Value range: 3.6 -
#> 12.8Highest values found in: Treatment BLowest values found in:
#> Treatment A
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ─────────────────────────────────────────
#> Statistic Value
#> ─────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 7.961667
#> Median Value 7.800000
#> Standard Deviation 2.004748
#> Value Range 3.6 - 12.8
#> Highest Value Group Treatment B
#> Lowest Value Group Treatment A
#> ─────────────────────────────────────────
#>
# Compare WBC across multiple factors
lollipop(
data = clinical_lab_data,
dep = "white_blood_cells",
group = "treatment_group",
conditionalColor = TRUE,
colorThreshold = 11, # Upper normal limit
orientation = "horizontal",
sortBy = "value_desc",
lineWidth = 2,
title = "WBC Count by Treatment (ULN: 11)"
)
#>
#> LOLLIPOP CHART
#>
#> Multiple observations per group detected (max=26 per group). Groups
#> with duplicates: Control, Treatment A, Treatment B. Use aggregation
#> (mean/median/sum) to avoid over-plotting and misleading visualization.
#>
#> Conditional coloring applied. Values > 11.00 colored orange (above
#> threshold), others blue (below).
#>
#> <div class='alert alert-success'>
#>
#> Clinical Summary
#>
#> Analysis Overview: This analysis compared 60 observations across 3
#> groups.
#>
#> Key Findings:
#>
#> Mean value: 7.96 (Standard Deviation = 2)Value range: 3.6 -
#> 12.8Highest values found in: Treatment BLowest values found in:
#> Treatment A
#>
#> Clinical Interpretation: Notable variation observed between groups,
#> suggesting clinically meaningful differences.
#>
#> Data Summary
#> ─────────────────────────────────────────
#> Statistic Value
#> ─────────────────────────────────────────
#> Number of Observations 60
#> Number of Groups 3
#> Mean Value 7.961667
#> Median Value 7.800000
#> Standard Deviation 2.004748
#> Value Range 3.6 - 12.8
#> Highest Value Group Treatment B
#> Lowest Value Group Treatment A
#> ─────────────────────────────────────────
#>