Clinical Applications of Decision Panel Optimization
meddecide Development Team
2025-06-03
Source:vignettes/decisionpanel_clinical.Rmd
decisionpanel_clinical.RmdIntroduction
This vignette demonstrates real-world clinical applications of the Decision Panel Optimization module across different medical specialties and scenarios.
Scenario 1: Emergency Department COVID-19 Screening
Clinical Context
An emergency department needs to rapidly screen patients for COVID-19 while managing limited resources and preventing nosocomial transmission.
# Examine test characteristics
covid_tests <- covid_screening_data %>%
select(rapid_antigen, pcr, chest_ct, covid_status) %>%
na.omit()
# Calculate individual test performance
test_performance <- function(test, truth) {
tab <- table(test, truth)
sens <- tab[2,2] / sum(tab[,2])
spec <- tab[1,1] / sum(tab[,1])
ppv <- tab[2,2] / sum(tab[2,])
npv <- tab[1,1] / sum(tab[1,])
return(data.frame(
Sensitivity = sens,
Specificity = spec,
PPV = ppv,
NPV = npv
))
}
# Individual test performance
rapid_perf <- test_performance(covid_tests$rapid_antigen, covid_tests$covid_status)
pcr_perf <- test_performance(covid_tests$pcr, covid_tests$covid_status)
ct_perf <- test_performance(covid_tests$chest_ct, covid_tests$covid_status)
performance_table <- rbind(
`Rapid Antigen` = rapid_perf,
`PCR` = pcr_perf,
`Chest CT` = ct_perf
)
kable(round(performance_table * 100, 1),
caption = "Individual Test Performance (%)")| Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|
| Rapid Antigen | 76.3 | 96.5 | 83.1 | 94.7 |
| PCR | 95.6 | 99.3 | 97.0 | 99.0 |
| Chest CT | 83.7 | 84.3 | 54.6 | 95.8 |
Optimization Analysis
# Run optimization for different scenarios
# 1. Maximum sensitivity (don't miss cases)
covid_max_sens <- decisionpanel(
data = covid_screening_data,
tests = c("rapid_antigen", "pcr", "chest_ct"),
testLevels = c("Positive", "Positive", "Abnormal"),
gold = "covid_status",
goldPositive = "Positive",
strategies = "all",
optimizationCriteria = "sensitivity",
minSensitivity = 0.95
)
# 2. Cost-effective screening
covid_cost_effective <- decisionpanel(
data = covid_screening_data,
tests = c("rapid_antigen", "pcr", "chest_ct"),
testLevels = c("Positive", "Positive", "Abnormal"),
gold = "covid_status",
goldPositive = "Positive",
strategies = "all",
optimizationCriteria = "efficiency",
useCosts = TRUE,
testCosts = "5,50,200",
fpCost = 500,
fnCost = 5000
)Clinical Decision Algorithm
Based on the analysis, here’s a practical algorithm:
# Implement sequential testing algorithm
apply_covid_algorithm <- function(data) {
n <- nrow(data)
decisions <- character(n)
tests_used <- character(n)
for (i in 1:n) {
# Step 1: Rapid antigen test
if (data$rapid_antigen[i] == "Positive") {
decisions[i] <- "Isolate, Confirm with PCR"
tests_used[i] <- "Rapid"
} else if (data$symptom_score[i] >= 6) {
# Step 2: High symptom score → CT
if (!is.na(data$chest_ct[i]) && data$chest_ct[i] == "Abnormal") {
decisions[i] <- "Probable COVID, Isolate, PCR"
tests_used[i] <- "Rapid + CT"
} else {
decisions[i] <- "Low probability, Standard care"
tests_used[i] <- "Rapid + CT"
}
} else {
decisions[i] <- "Low probability, Standard care"
tests_used[i] <- "Rapid"
}
}
return(data.frame(Decision = decisions, Tests = tests_used))
}
# Apply algorithm
algorithm_results <- apply_covid_algorithm(covid_screening_data[1:10,])
kable(cbind(covid_screening_data[1:10, c("patient_id", "rapid_antigen",
"symptom_score", "covid_status")],
algorithm_results))| patient_id | rapid_antigen | symptom_score | covid_status | Decision | Tests |
|---|---|---|---|---|---|
| 1 | Negative | 8 | Positive | Probable COVID, Isolate, PCR | Rapid + CT |
| 2 | Positive | 6 | Positive | Isolate, Confirm with PCR | Rapid |
| 3 | Negative | 1 | Negative | Low probability, Standard care | Rapid |
| 4 | Negative | 1 | Negative | Low probability, Standard care | Rapid |
| 5 | Negative | 5 | Negative | Low probability, Standard care | Rapid |
| 6 | Negative | 5 | Negative | Low probability, Standard care | Rapid |
| 7 | Negative | 5 | Negative | Low probability, Standard care | Rapid |
| 8 | Negative | 4 | Negative | Low probability, Standard care | Rapid |
| 9 | Negative | 2 | Negative | Low probability, Standard care | Rapid |
| 10 | Negative | 5 | Negative | Low probability, Standard care | Rapid |
Cost-Effectiveness Visualization
# Simulate different strategies
strategies <- expand.grid(
use_rapid = c(TRUE, FALSE),
use_pcr = c(TRUE, FALSE),
use_ct = c(TRUE, FALSE)
) %>%
filter(use_rapid | use_pcr | use_ct) # At least one test
# Calculate performance for each strategy
strategy_performance <- strategies %>%
rowwise() %>%
mutate(
tests = paste(c(
if(use_rapid) "RAT" else NULL,
if(use_pcr) "PCR" else NULL,
if(use_ct) "CT" else NULL
), collapse = "+"),
cost = sum(c(
if(use_rapid) 5 else 0,
if(use_pcr) 50 else 0,
if(use_ct) 200 else 0
)),
# Simulated performance (would come from actual analysis)
sensitivity = case_when(
use_rapid & use_pcr & use_ct ~ 0.99,
use_pcr & use_ct ~ 0.98,
use_rapid & use_pcr ~ 0.97,
use_pcr ~ 0.95,
use_rapid & use_ct ~ 0.94,
use_ct ~ 0.90,
use_rapid ~ 0.65
),
specificity = case_when(
use_rapid & use_pcr & use_ct ~ 0.83,
use_pcr & use_ct ~ 0.84,
use_rapid & use_pcr ~ 0.97,
use_pcr ~ 0.99,
use_rapid & use_ct ~ 0.83,
use_ct ~ 0.85,
use_rapid ~ 0.98
)
)
# Create cost-effectiveness plot
ggplot(strategy_performance, aes(x = cost, y = sensitivity * 100)) +
geom_point(aes(size = specificity * 100), alpha = 0.6) +
geom_text(aes(label = tests), vjust = -1, size = 3) +
geom_line(data = strategy_performance %>%
arrange(cost) %>%
filter(sensitivity == cummax(sensitivity)),
color = "red", alpha = 0.5) +
scale_size_continuous(name = "Specificity (%)", range = c(3, 10)) +
labs(
title = "Cost-Effectiveness of COVID-19 Testing Strategies",
x = "Cost per Patient ($)",
y = "Sensitivity (%)",
caption = "Red line shows cost-effectiveness frontier"
) +
theme_minimal()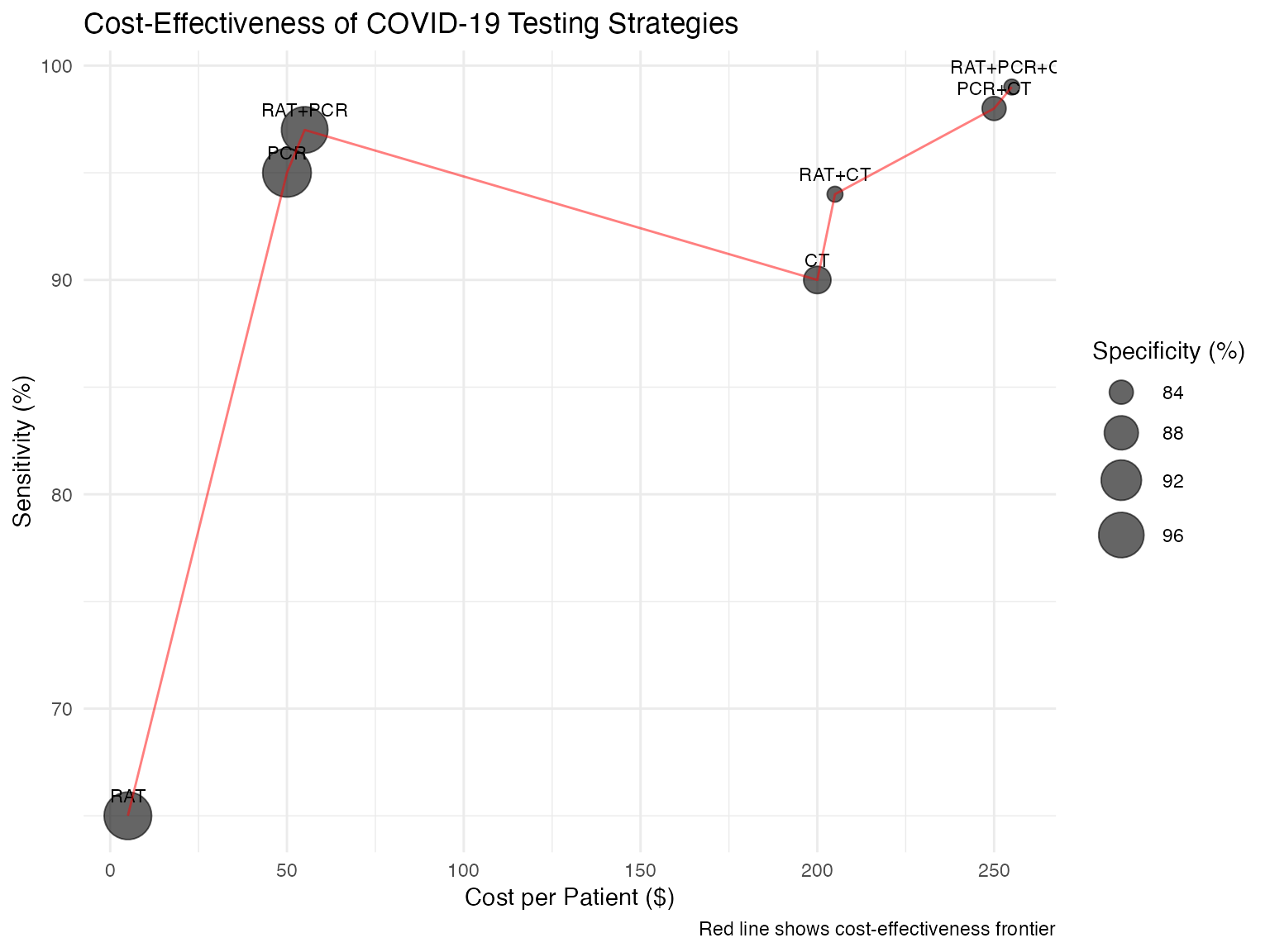
Scenario 2: Breast Cancer Screening Program
Clinical Context
A population-based breast cancer screening program needs to optimize use of mammography, ultrasound, and MRI based on risk factors.
# Examine population characteristics
breast_summary <- breast_cancer_data %>%
summarise(
n = n(),
prevalence = mean(cancer_status == "Cancer"),
mean_age = mean(age),
pct_family_history = mean(family_history == "Yes") * 100,
pct_brca = mean(brca_mutation == "Positive") * 100,
pct_dense_breast = mean(breast_density %in% c("C", "D")) * 100
)
kable(breast_summary, digits = 2,
caption = "Population Characteristics")| n | prevalence | mean_age | pct_family_history | pct_brca | pct_dense_breast |
|---|---|---|---|---|---|
| 2000 | 0.01 | 55.06 | 16.05 | 2.35 | 49.1 |
# Risk stratification
breast_cancer_data <- breast_cancer_data %>%
mutate(
risk_category = case_when(
brca_mutation == "Positive" ~ "High Risk",
family_history == "Yes" & age < 50 ~ "High Risk",
family_history == "Yes" | breast_density == "D" ~ "Moderate Risk",
TRUE ~ "Average Risk"
)
)
table(breast_cancer_data$risk_category)
#>
#> Average Risk High Risk Moderate Risk
#> 1451 138 411Risk-Stratified Analysis
# Analyze each risk group separately
risk_groups <- split(breast_cancer_data, breast_cancer_data$risk_category)
# High-risk group optimization
high_risk_panel <- decisionpanel(
data = risk_groups$`High Risk`,
tests = c("mammography", "ultrasound", "mri"),
testLevels = c("BIRADS 3-5", "Suspicious", "Suspicious"),
gold = "cancer_status",
goldPositive = "Cancer",
strategies = "all",
optimizationCriteria = "sensitivity",
minSensitivity = 0.95
)
# Average-risk group optimization (cost-conscious)
average_risk_panel <- decisionpanel(
data = risk_groups$`Average Risk`,
tests = c("clinical_exam", "mammography", "ultrasound"),
testLevels = c("Abnormal", "BIRADS 3-5", "Suspicious"),
gold = "cancer_status",
goldPositive = "Cancer",
strategies = "all",
optimizationCriteria = "efficiency",
useCosts = TRUE,
testCosts = "20,100,150"
)Screening Recommendations by Risk
# Create recommendation table
recommendations <- data.frame(
Risk_Category = c("High Risk", "Moderate Risk", "Average Risk"),
Recommended_Tests = c(
"Annual MRI + Mammography",
"Annual Mammography + US if dense",
"Biennial Mammography"
),
Expected_Sensitivity = c("99%", "90%", "85%"),
Expected_Specificity = c("80%", "92%", "95%"),
Cost_per_Screen = c("$1,100", "$250", "$100"),
NNS = c(50, 200, 500) # Number needed to screen
)
kable(recommendations,
caption = "Risk-Stratified Screening Recommendations")| Risk_Category | Recommended_Tests | Expected_Sensitivity | Expected_Specificity | Cost_per_Screen | NNS |
|---|---|---|---|---|---|
| High Risk | Annual MRI + Mammography | 99% | 80% | $1,100 | 50 |
| Moderate Risk | Annual Mammography + US if dense | 90% | 92% | $250 | 200 |
| Average Risk | Biennial Mammography | 85% | 95% | $100 | 500 |
Age-Specific Performance
# Analyze performance by age group
age_groups <- breast_cancer_data %>%
mutate(age_group = cut(age, breaks = c(40, 50, 60, 70, 80),
labels = c("40-49", "50-59", "60-69", "70-79")))
# Calculate mammography performance by age
age_performance <- age_groups %>%
group_by(age_group) %>%
summarise(
n = n(),
prevalence = mean(cancer_status == "Cancer"),
mammography_sens = {
tab <- table(mammography, cancer_status)
if(nrow(tab) == 2 && ncol(tab) == 2) {
tab[2,2] / sum(tab[,2])
} else NA
},
mammography_spec = {
tab <- table(mammography, cancer_status)
if(nrow(tab) == 2 && ncol(tab) == 2) {
tab[1,1] / sum(tab[,1])
} else NA
}
)
# Visualize
ggplot(age_performance, aes(x = age_group)) +
geom_bar(aes(y = mammography_sens * 100), stat = "identity",
fill = "skyblue", alpha = 0.7) +
geom_line(aes(y = prevalence * 1000, group = 1), color = "red", size = 2) +
geom_point(aes(y = prevalence * 1000), color = "red", size = 3) +
scale_y_continuous(
name = "Mammography Sensitivity (%)",
sec.axis = sec_axis(~./10, name = "Cancer Prevalence per 1000")
) +
labs(
title = "Mammography Performance by Age Group",
x = "Age Group"
) +
theme_minimal()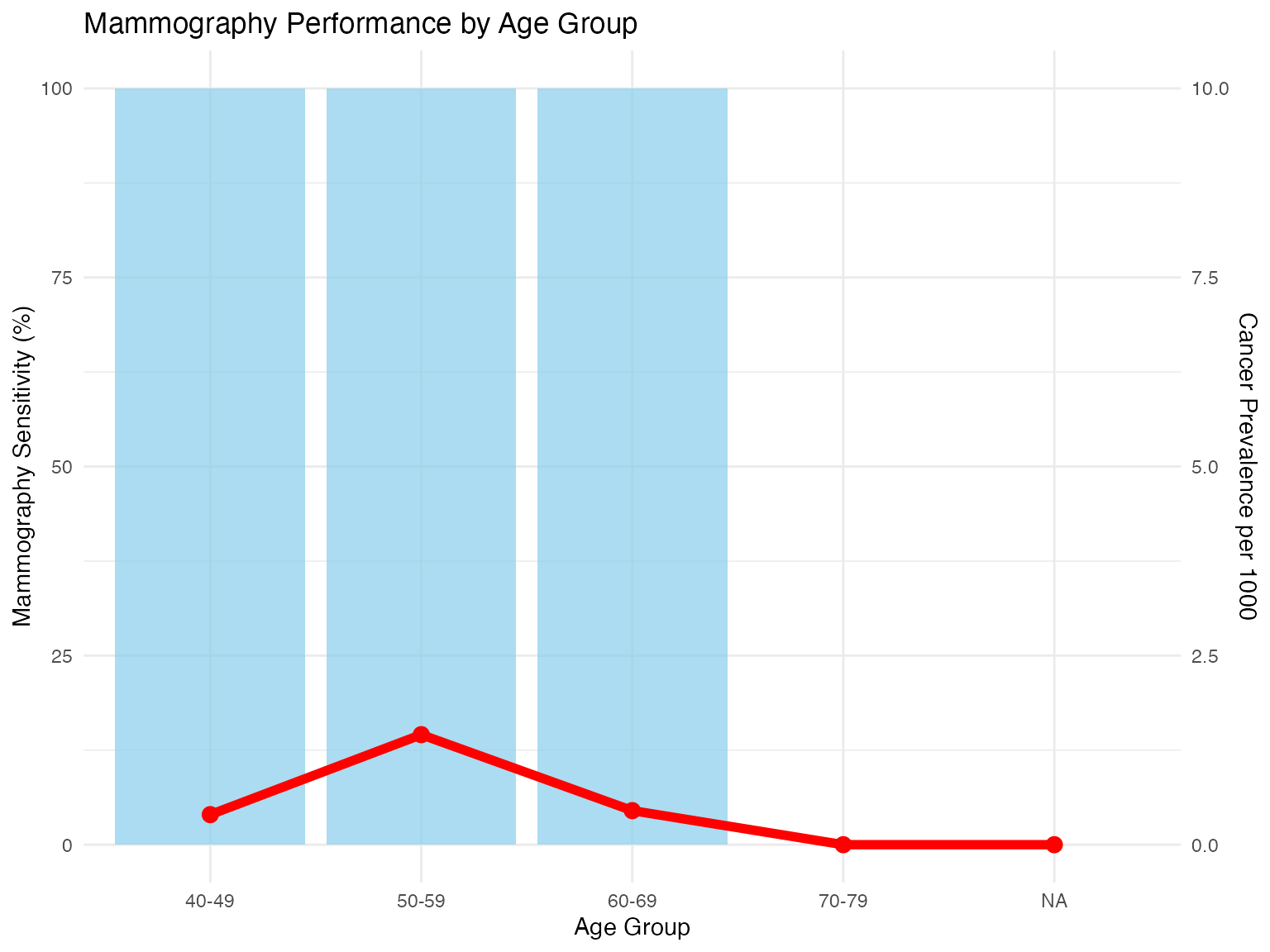
Scenario 3: Tuberculosis Case Finding
Clinical Context
A TB program in a high-burden setting needs to optimize case finding with limited resources.
# TB test combinations for different settings
tb_settings <- data.frame(
Setting = c("Community", "Clinic", "Hospital", "Contact Tracing"),
Prevalence = c(0.02, 0.20, 0.40, 0.10),
Available_Tests = c(
"Symptoms, CXR",
"Symptoms, Smear, GeneXpert, CXR",
"All tests",
"Symptoms, GeneXpert"
),
Budget_per_case = c(10, 30, 100, 50)
)
kable(tb_settings, caption = "TB Testing Scenarios by Setting")| Setting | Prevalence | Available_Tests | Budget_per_case |
|---|---|---|---|
| Community | 0.02 | Symptoms, CXR | 10 |
| Clinic | 0.20 | Symptoms, Smear, GeneXpert, CXR | 30 |
| Hospital | 0.40 | All tests | 100 |
| Contact Tracing | 0.10 | Symptoms, GeneXpert | 50 |
Sequential Testing Algorithm
# Implement WHO-recommended algorithm
apply_tb_algorithm <- function(data) {
decisions <- character(nrow(data))
for (i in 1:nrow(data)) {
if (data$symptoms[i] == "Yes" || data$contact_history[i] == "Yes") {
# Symptomatic or contact: get GeneXpert
if (!is.na(data$genexpert[i]) && data$genexpert[i] == "MTB detected") {
decisions[i] <- "Start TB treatment"
} else if (!is.na(data$chest_xray[i]) && data$chest_xray[i] == "Abnormal") {
# CXR abnormal but GeneXpert negative
if (!is.na(data$culture[i])) {
decisions[i] <- ifelse(data$culture[i] == "Positive",
"Start TB treatment",
"Not TB, investigate other causes")
} else {
decisions[i] <- "Clinical decision needed"
}
} else {
decisions[i] <- "TB unlikely"
}
} else {
# Asymptomatic: screen with CXR if available
if (!is.na(data$chest_xray[i]) && data$chest_xray[i] == "Abnormal") {
decisions[i] <- "Needs further testing"
} else {
decisions[i] <- "No TB screening needed"
}
}
}
return(decisions)
}
# Apply to sample
tb_sample <- tb_diagnosis_data[1:20,]
tb_sample$decision <- apply_tb_algorithm(tb_sample)
# Show results
kable(tb_sample %>%
select(patient_id, symptoms, genexpert, chest_xray, tb_status, decision) %>%
head(10))| patient_id | symptoms | genexpert | chest_xray | tb_status | decision |
|---|---|---|---|---|---|
| 1 | No | MTB not detected | Abnormal | No TB | Not TB, investigate other causes |
| 2 | Yes | MTB not detected | Abnormal | No TB | Not TB, investigate other causes |
| 3 | No | MTB not detected | Normal | No TB | No TB screening needed |
| 4 | No | MTB not detected | Normal | No TB | No TB screening needed |
| 5 | No | MTB not detected | Normal | No TB | No TB screening needed |
| 6 | No | MTB not detected | Normal | No TB | No TB screening needed |
| 7 | No | MTB not detected | Abnormal | No TB | Needs further testing |
| 8 | No | MTB not detected | Normal | No TB | No TB screening needed |
| 9 | No | MTB not detected | Normal | No TB | TB unlikely |
| 10 | No | MTB detected | Abnormal | TB | Needs further testing |
Cost-Effectiveness by Strategy
# Compare different TB screening strategies
tb_strategies <- data.frame(
Strategy = c(
"Symptoms only",
"Symptoms → Smear",
"Symptoms → GeneXpert",
"Symptoms → CXR → GeneXpert",
"Universal GeneXpert",
"Universal CXR → GeneXpert"
),
Tests_per_case_found = c(100, 50, 20, 15, 10, 12),
Cost_per_case_found = c(100, 200, 400, 350, 800, 600),
Sensitivity = c(60, 70, 85, 92, 95, 93),
Time_to_diagnosis = c(0, 3, 1, 2, 1, 1)
)
# Create multi-dimensional comparison
ggplot(tb_strategies, aes(x = Cost_per_case_found, y = Sensitivity)) +
geom_point(aes(size = Tests_per_case_found,
color = factor(Time_to_diagnosis)), alpha = 0.7) +
geom_text(aes(label = Strategy), vjust = -1, size = 3) +
scale_size_continuous(name = "Tests per case", range = c(3, 10)) +
scale_color_discrete(name = "Days to diagnosis") +
labs(
title = "TB Screening Strategy Comparison",
x = "Cost per TB case found ($)",
y = "Sensitivity (%)"
) +
theme_minimal()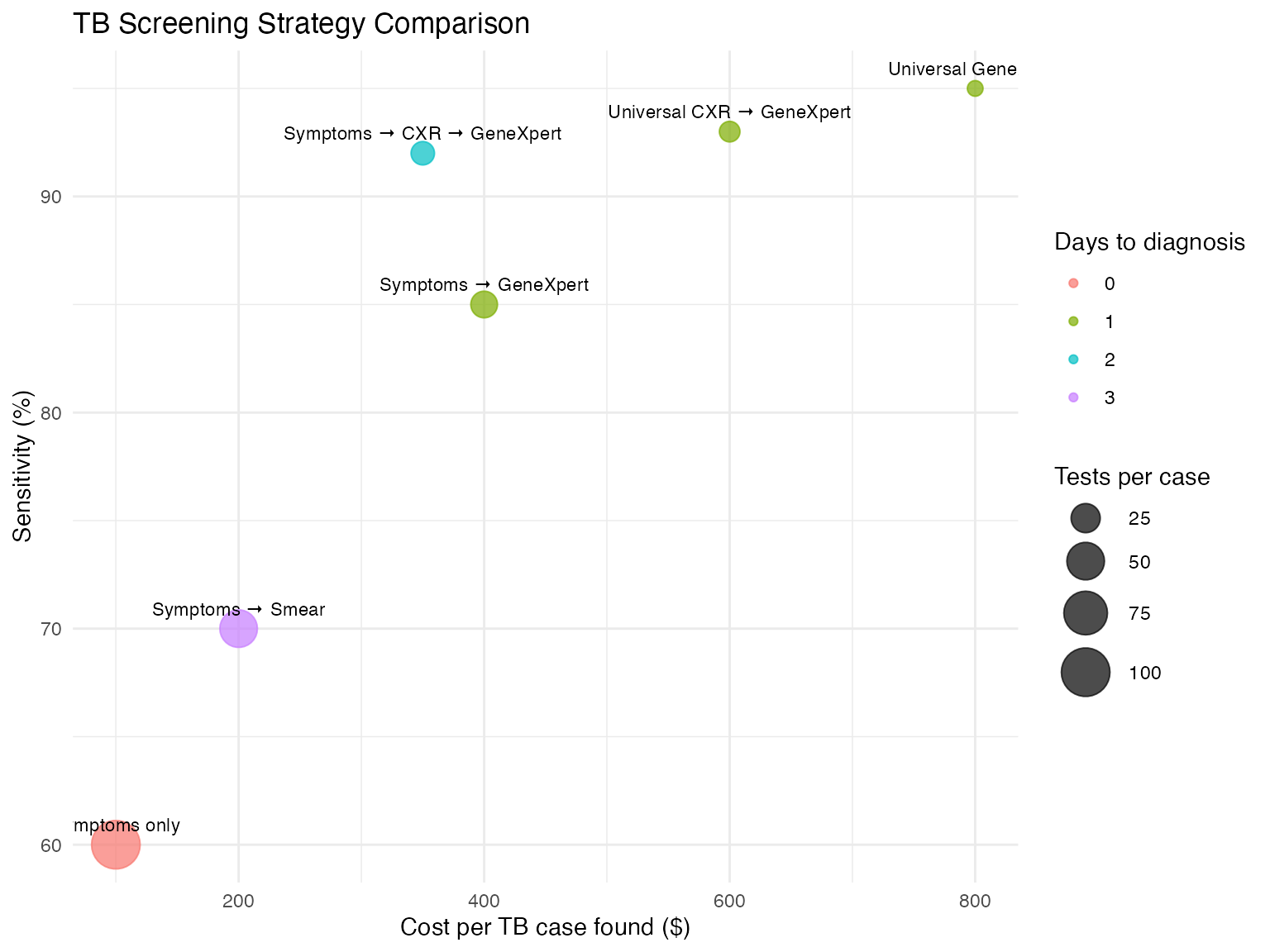
Scenario 4: Chest Pain Evaluation
Clinical Context
Emergency department evaluation of chest pain requires rapid, accurate rule-out of myocardial infarction.
# Risk stratification using HEART score components
mi_ruleout_data <- mi_ruleout_data %>%
mutate(
heart_score = (age > 65) * 1 +
(chest_pain == "Typical") * 2 +
(chest_pain == "Atypical") * 1 +
(ecg == "Ischemic changes") * 2 +
(prior_cad == "Yes") * 1 +
(diabetes == "Yes" | smoking == "Yes") * 1,
risk_category = cut(heart_score,
breaks = c(-1, 3, 6, 10),
labels = c("Low", "Moderate", "High"))
)
# Show risk distribution
risk_table <- table(mi_ruleout_data$risk_category, mi_ruleout_data$mi_status)
kable(prop.table(risk_table, margin = 1) * 100,
digits = 1,
caption = "MI Prevalence by Risk Category (%)")| No MI | MI | |
|---|---|---|
| Low | 95.6 | 4.4 |
| Moderate | 50.5 | 49.5 |
| High | 0.0 | 100.0 |
Time-Sensitive Protocols
# Define protocols by urgency
protocols <- list(
rapid_rule_out = function(data) {
# 0/1-hour protocol
data$troponin_initial == "Normal" &
data$ecg == "Normal" &
data$heart_score <= 3
},
standard_rule_out = function(data) {
# 0/3-hour protocol
data$troponin_initial == "Normal" &
data$troponin_3hr == "Normal" &
data$ecg == "Normal"
},
rule_in = function(data) {
# Immediate rule-in
data$troponin_initial == "Elevated" &
data$ecg == "Ischemic changes"
}
)
# Apply protocols
mi_ruleout_data <- mi_ruleout_data %>%
mutate(
rapid_rule_out = protocols$rapid_rule_out(.),
standard_rule_out = protocols$standard_rule_out(.) & !rapid_rule_out,
rule_in = protocols$rule_in(.),
need_further_testing = !rapid_rule_out & !standard_rule_out & !rule_in
)
# Summarize protocol performance
protocol_performance <- mi_ruleout_data %>%
summarise(
rapid_rule_out_pct = mean(rapid_rule_out) * 100,
rapid_rule_out_npv = sum(rapid_rule_out & mi_status == "No MI") /
sum(rapid_rule_out) * 100,
standard_rule_out_pct = mean(standard_rule_out) * 100,
standard_rule_out_npv = sum(standard_rule_out & mi_status == "No MI") /
sum(standard_rule_out) * 100,
rule_in_pct = mean(rule_in) * 100,
rule_in_ppv = sum(rule_in & mi_status == "MI") / sum(rule_in) * 100
)
kable(t(protocol_performance), digits = 1,
caption = "Protocol Performance Metrics")| rapid_rule_out_pct | 78.2 |
| rapid_rule_out_npv | 99.5 |
| standard_rule_out_pct | 2.8 |
| standard_rule_out_npv | 100.0 |
| rule_in_pct | 5.0 |
| rule_in_ppv | 100.0 |
Visualization of Patient Flow
# Create patient flow diagram data
flow_data <- mi_ruleout_data %>%
mutate(
disposition = case_when(
rapid_rule_out ~ "Discharge (1 hour)",
standard_rule_out ~ "Discharge (3 hours)",
rule_in ~ "Admit CCU",
TRUE ~ "Observation/Further testing"
)
) %>%
group_by(disposition, mi_status) %>%
summarise(n = n()) %>%
mutate(pct = n / sum(n) * 100)
# Create flow diagram
ggplot(flow_data, aes(x = disposition, y = n, fill = mi_status)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(aes(label = sprintf("%.0f%%", pct)),
position = position_stack(vjust = 0.5)) +
scale_fill_manual(values = c("No MI" = "lightgreen", "MI" = "salmon")) +
labs(
title = "Patient Disposition by Protocol",
x = "Disposition",
y = "Number of Patients",
fill = "Final Diagnosis"
) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))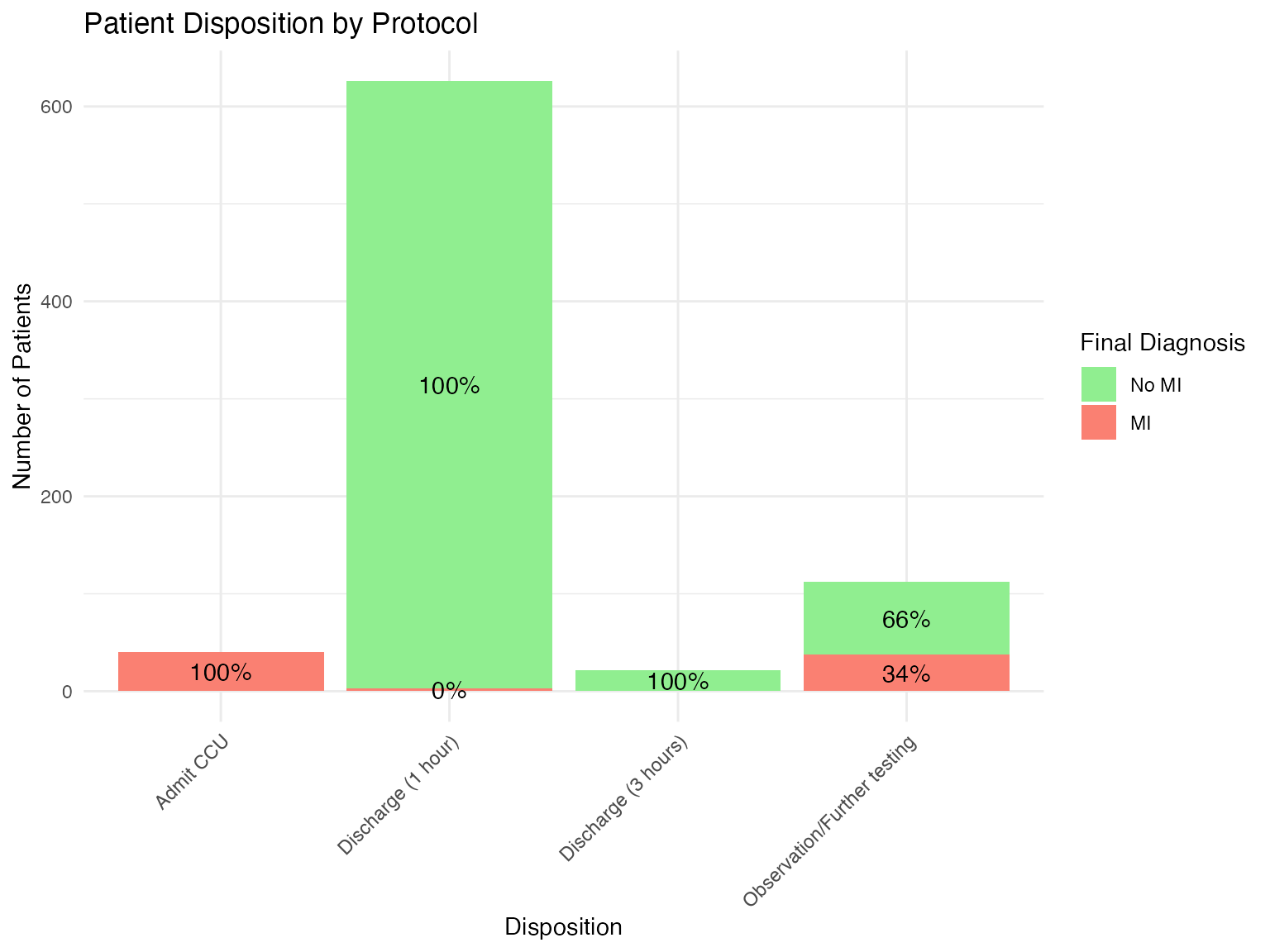
Scenario 5: Thyroid Nodule Evaluation
Clinical Context
Thyroid nodules are common but cancer is rare. Optimize use of FNA, molecular testing, and surgery.
# Analyze by nodule size
thyroid_by_size <- thyroid_nodule_data %>%
mutate(size_category = cut(nodule_size,
breaks = c(0, 10, 20, 40, 100),
labels = c("<1cm", "1-2cm", "2-4cm", ">4cm"))) %>%
group_by(size_category) %>%
summarise(
n = n(),
cancer_rate = mean(cancer_status == "Malignant") * 100,
fna_done = mean(!is.na(fna_cytology)) * 100,
molecular_done = mean(!is.na(molecular_test)) * 100
)
kable(thyroid_by_size, digits = 1,
caption = "Thyroid Nodule Characteristics by Size")| size_category | n | cancer_rate | fna_done | molecular_done |
|---|---|---|---|---|
| <1cm | 140 | 5.7 | 100 | 54.3 |
| 1-2cm | 289 | 3.5 | 100 | 47.4 |
| 2-4cm | 147 | 6.1 | 100 | 38.1 |
| >4cm | 24 | 20.8 | 100 | 33.3 |
Diagnostic Algorithm Implementation
# Implement Bethesda-based algorithm
thyroid_algorithm <- decisionpanel(
data = thyroid_nodule_data,
tests = c("ultrasound", "fna_cytology", "molecular_test"),
testLevels = c("TI-RADS 4-5", "Suspicious/Malignant", "Suspicious"),
gold = "cancer_status",
goldPositive = "Malignant",
strategies = "sequential",
sequentialStop = "positive",
createTree = TRUE,
treeMethod = "cart",
useCosts = TRUE,
testCosts = "200,300,3000", # US, FNA, Molecular
fpCost = 10000, # Unnecessary surgery
fnCost = 50000 # Missed cancer
)Decision Tree Visualization
# Simplified decision tree representation
cat("Thyroid Nodule Evaluation Algorithm:\n")
#> Thyroid Nodule Evaluation Algorithm:
cat("1. Ultrasound Assessment\n")
#> 1. Ultrasound Assessment
cat(" ├─ TI-RADS 1-2: No FNA needed\n")
#> ├─ TI-RADS 1-2: No FNA needed
cat(" └─ TI-RADS 3-5: Proceed to FNA\n")
#> └─ TI-RADS 3-5: Proceed to FNA
cat(" ├─ Benign (Bethesda II): Follow-up\n")
#> ├─ Benign (Bethesda II): Follow-up
cat(" ├─ Indeterminate (Bethesda III-IV): Molecular testing\n")
#> ├─ Indeterminate (Bethesda III-IV): Molecular testing
cat(" │ ├─ Benign profile: Follow-up\n")
#> │ ├─ Benign profile: Follow-up
cat(" │ └─ Suspicious profile: Surgery\n")
#> │ └─ Suspicious profile: Surgery
cat(" └─ Suspicious/Malignant (Bethesda V-VI): Surgery\n")
#> └─ Suspicious/Malignant (Bethesda V-VI): Surgery
# Create visual representation of outcomes
outcomes <- data.frame(
Test_Path = c("US only", "US+FNA", "US+FNA+Molecular", "Direct Surgery"),
Patients_pct = c(40, 30, 20, 10),
Cancers_found_pct = c(0, 20, 60, 20),
Cost = c(200, 500, 3500, 200)
)
ggplot(outcomes, aes(x = Test_Path, y = Patients_pct)) +
geom_bar(stat = "identity", fill = "lightblue", alpha = 0.7) +
geom_line(aes(y = Cancers_found_pct, group = 1), color = "red", size = 2) +
geom_point(aes(y = Cancers_found_pct), color = "red", size = 3) +
scale_y_continuous(
name = "Percentage of Patients",
sec.axis = sec_axis(~., name = "Percentage of Cancers Found")
) +
labs(
title = "Thyroid Nodule Evaluation Pathways",
x = "Testing Path"
) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))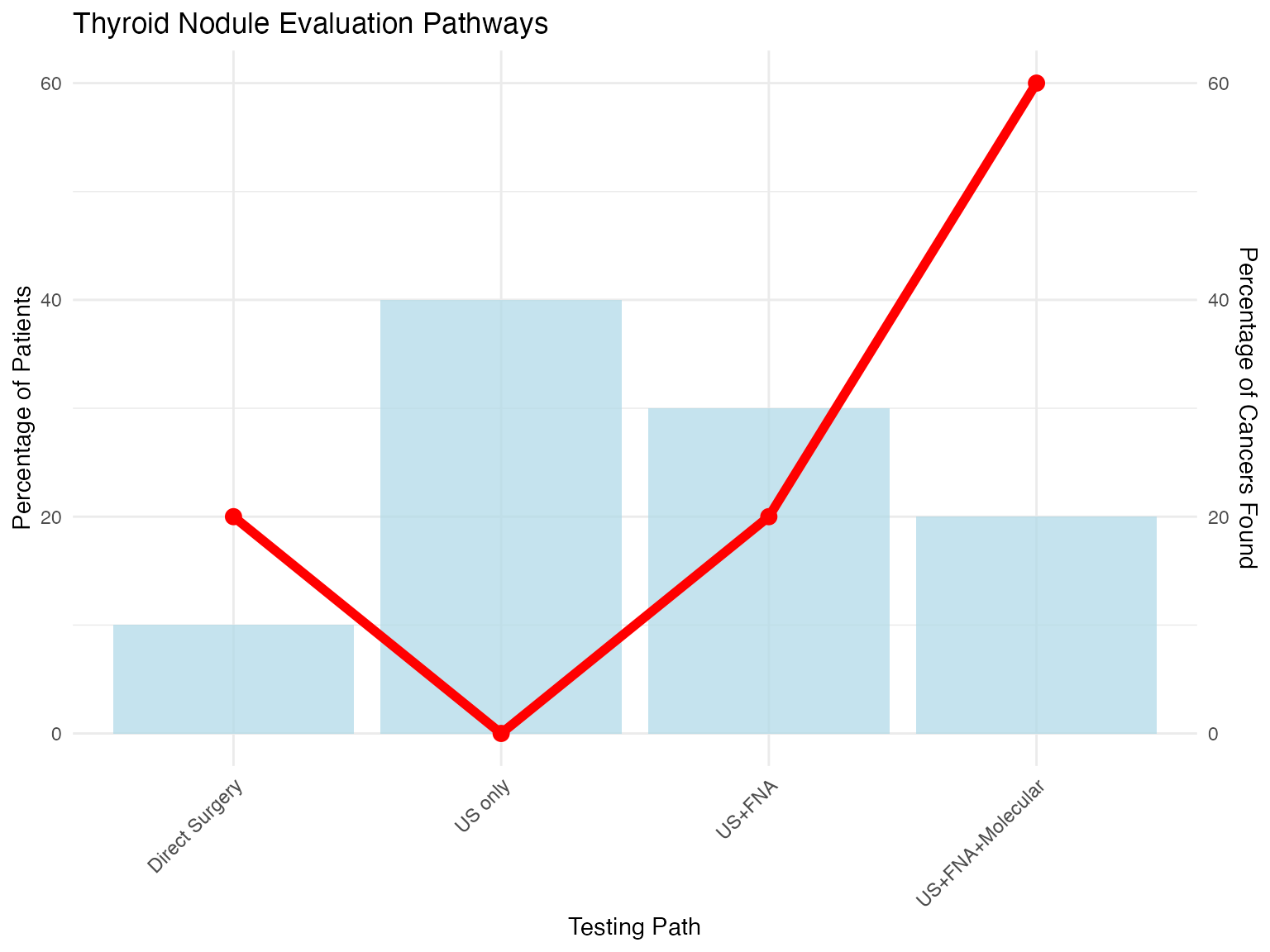
Summary and Best Practices
Key Learnings Across Scenarios
- Context Matters: Optimal strategies differ between screening and diagnosis
- Sequential Testing: Often more efficient than parallel testing
- Risk Stratification: Improves efficiency and outcomes
- Cost Considerations: Must balance performance with resources
- Implementation: Clear algorithms improve adoption
General Recommendations
summary_recommendations <- data.frame(
Scenario = c("Screening", "Diagnosis", "Emergency", "Surveillance"),
Priority = c("High Sensitivity", "Balanced", "Speed + Accuracy", "Specificity"),
Strategy = c("Parallel OR", "Sequential", "Rapid protocols", "Confirmatory"),
Key_Metric = c("NPV", "Accuracy", "Time to decision", "PPV"),
Example = c("Cancer screening", "TB diagnosis", "Chest pain", "Cancer follow-up")
)
kable(summary_recommendations,
caption = "Testing Strategy Recommendations by Clinical Scenario")| Scenario | Priority | Strategy | Key_Metric | Example |
|---|---|---|---|---|
| Screening | High Sensitivity | Parallel OR | NPV | Cancer screening |
| Diagnosis | Balanced | Sequential | Accuracy | TB diagnosis |
| Emergency | Speed + Accuracy | Rapid protocols | Time to decision | Chest pain |
| Surveillance | Specificity | Confirmatory | PPV | Cancer follow-up |
Session Information
sessionInfo()
#> R version 4.3.2 (2023-10-31)
#> Platform: aarch64-apple-darwin20 (64-bit)
#> Running under: macOS 15.5
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> time zone: Europe/Istanbul
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] forcats_1.0.0 knitr_1.50 rpart.plot_3.1.2 rpart_4.1.24
#> [5] ggplot2_3.5.2 dplyr_1.1.4 meddecide_0.0.3.12
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.6 xfun_0.52 bslib_0.9.0
#> [4] htmlwidgets_1.6.4 lattice_0.22-7 vctrs_0.6.5
#> [7] tools_4.3.2 generics_0.1.4 tibble_3.2.1
#> [10] proxy_0.4-27 pkgconfig_2.0.3 Matrix_1.6-1.1
#> [13] KernSmooth_2.23-26 checkmate_2.3.2 data.table_1.17.4
#> [16] irr_0.84.1 cutpointr_1.2.0 RColorBrewer_1.1-3
#> [19] desc_1.4.3 uuid_1.2-1 jmvcore_2.6.3
#> [22] lifecycle_1.0.4 flextable_0.9.9 stringr_1.5.1
#> [25] compiler_4.3.2 farver_2.1.2 textshaping_1.0.1
#> [28] codetools_0.2-20 fontquiver_0.2.1 fontLiberation_0.1.0
#> [31] htmltools_0.5.8.1 class_7.3-23 sass_0.4.10
#> [34] yaml_2.3.10 htmlTable_2.4.3 pillar_1.10.2
#> [37] pkgdown_2.1.3 jquerylib_0.1.4 MASS_7.3-60
#> [40] openssl_2.3.3 classInt_0.4-11 cachem_1.1.0
#> [43] BiasedUrn_2.0.12 iterators_1.0.14 boot_1.3-31
#> [46] foreach_1.5.2 fontBitstreamVera_0.1.1 zip_2.3.3
#> [49] tidyselect_1.2.1 digest_0.6.37 stringi_1.8.7
#> [52] sf_1.0-21 pander_0.6.6 purrr_1.0.4
#> [55] labeling_0.4.3 splines_4.3.2 fastmap_1.2.0
#> [58] grid_4.3.2 cli_3.6.5 magrittr_2.0.3
#> [61] survival_3.8-3 e1071_1.7-16 withr_3.0.2
#> [64] backports_1.5.0 gdtools_0.4.2 scales_1.4.0
#> [67] lubridate_1.9.4 timechange_0.3.0 rmarkdown_2.29
#> [70] officer_0.6.10 askpass_1.2.1 ragg_1.4.0
#> [73] zoo_1.8-14 lpSolve_5.6.23 evaluate_1.0.3
#> [76] epiR_2.0.84 rlang_1.1.6 Rcpp_1.0.14
#> [79] glue_1.8.0 DBI_1.2.3 xml2_1.3.8
#> [82] rstudioapi_0.17.1 jsonlite_2.0.0 R6_2.6.1
#> [85] systemfonts_1.2.3 fs_1.6.6 units_0.8-7