# For vignette demonstration, ensure 'histopathology' is available.
if (!exists("histopathology")) {
set.seed(123)
histopathology <- data.frame(
LVI = factor(sample(c("Present", "Absent", NA), 150, replace = TRUE, prob = c(0.3, 0.6, 0.1))),
PNI = factor(sample(c("Present", "Absent"), 150, replace = TRUE, prob = c(0.2, 0.8))),
PreinvasiveComponent = factor(sample(c("Yes", "No"), 150, replace = TRUE, prob = c(0.4, 0.6)))
)
}
head(histopathology)
#> LVI PNI PreinvasiveComponent
#> 1 Absent Present Yes
#> 2 Present Absent No
#> 3 Absent Absent Yes
#> 4 Present Absent Yes
#> 5 <NA> Absent Yes
#> 6 Absent Absent No
library(ClinicoPath)
#> Registered S3 method overwritten by 'future':
#> method from
#> all.equal.connection parallelly
#> Warning: replacing previous import 'dplyr::select' by 'jmvcore::select' when
#> loading 'ClinicoPath'
#> Warning: replacing previous import 'cutpointr::roc' by 'pROC::roc' when loading
#> 'ClinicoPath'
#> Warning: replacing previous import 'cutpointr::auc' by 'pROC::auc' when loading
#> 'ClinicoPath'
#> Warning: replacing previous import 'magrittr::extract' by 'tidyr::extract' when
#> loading 'ClinicoPath'
#> Warning in check_dep_version(): ABI version mismatch:
#> lme4 was built with Matrix ABI version 1
#> Current Matrix ABI version is 0
#> Please re-install lme4 from source or restore original 'Matrix' package
#> Warning: replacing previous import 'jmvcore::select' by 'dplyr::select' when
#> loading 'ClinicoPath'
#> Registered S3 methods overwritten by 'ggpp':
#> method from
#> heightDetails.titleGrob ggplot2
#> widthDetails.titleGrob ggplot2
#> Warning: replacing previous import 'DataExplorer::plot_histogram' by
#> 'grafify::plot_histogram' when loading 'ClinicoPath'
#> Warning: replacing previous import 'ROCR::plot' by 'graphics::plot' when
#> loading 'ClinicoPath'
#> Warning: replacing previous import 'dplyr::select' by 'jmvcore::select' when
#> loading 'ClinicoPath'
#> Warning: replacing previous import 'tibble::view' by 'summarytools::view' when
#> loading 'ClinicoPath'
#>
#> Attaching package: 'ClinicoPath'
#> The following object is masked _by_ '.GlobalEnv':
#>
#> histopathology
# data(histopathology, package="ClinicoPathDescriptives") # if needed
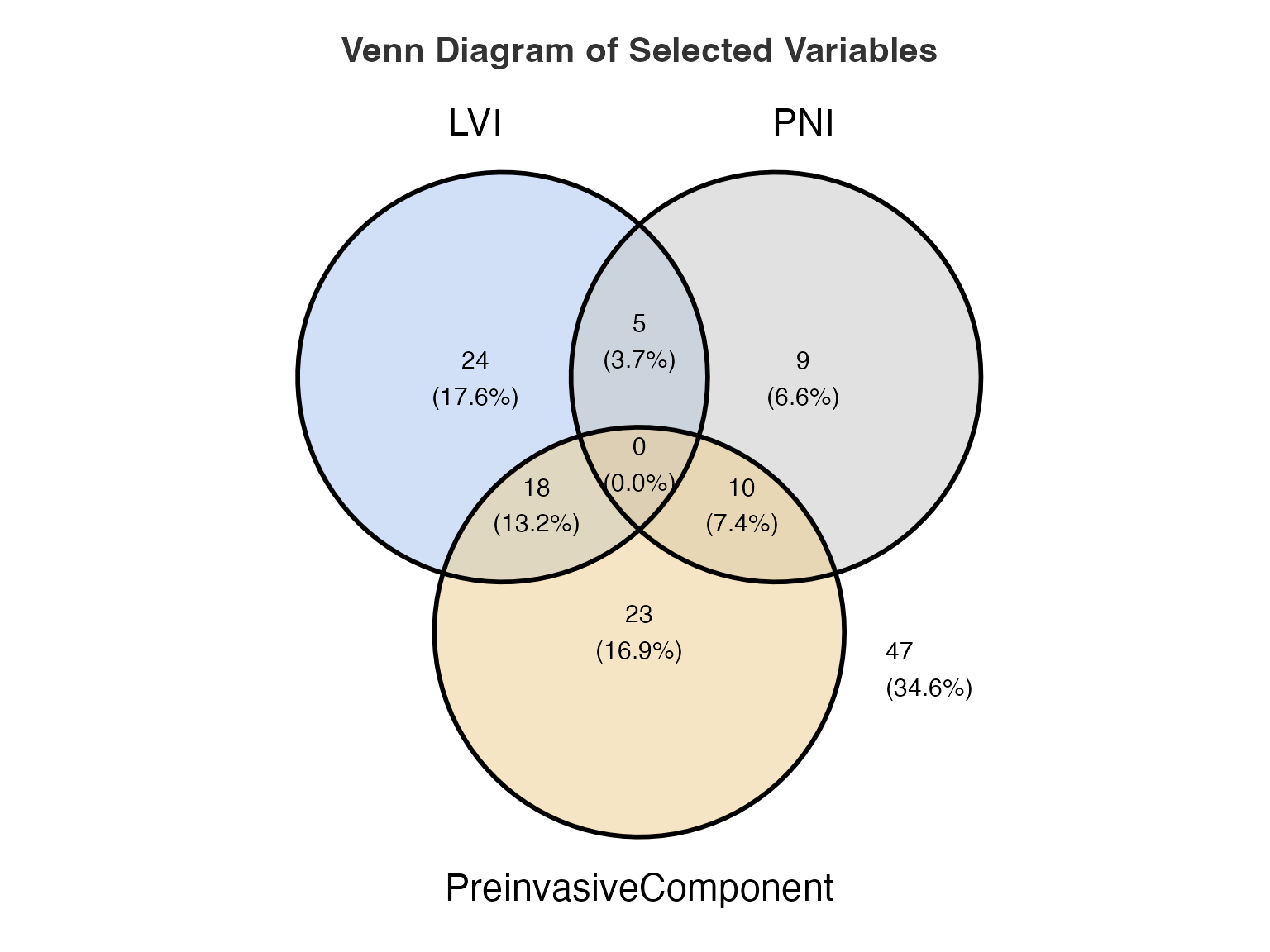
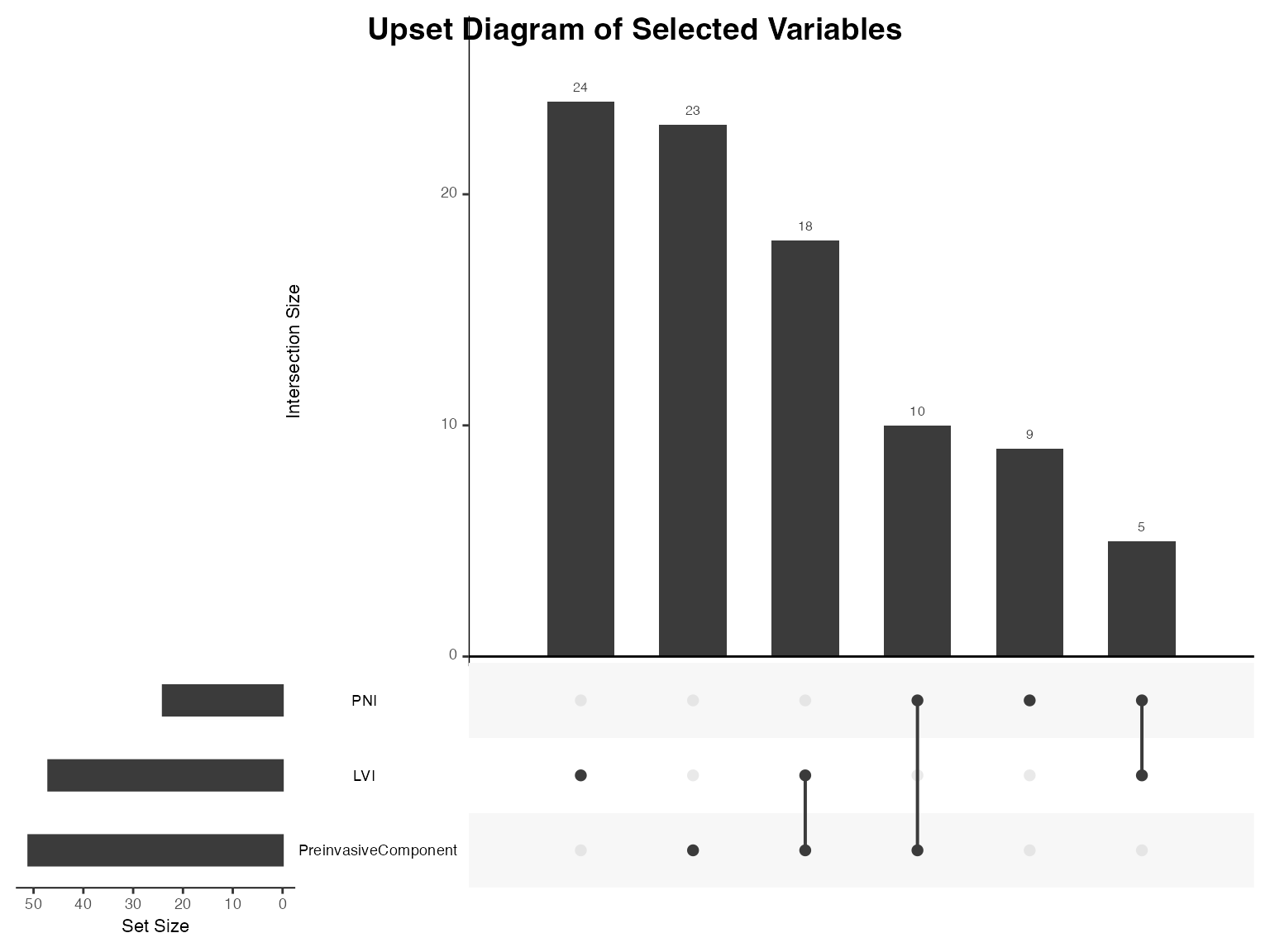
results_venn <- ClinicoPath::venn(
data = histopathology,
var1 = "LVI",
var1true = "Present",
var2 = "PNI",
var2true = "Present",
var3 = "PreinvasiveComponent",
var3true = "Yes",
var4 = NULL, # Explicitly NULL for unused variable
var4true = NULL # Explicitly NULL for unused true level
)