# For vignette demonstration, ensure 'treatmentResponse' is available.
# This data is part of the ClinicoPathDescriptives package.
data(treatmentResponse, package = "ClinicoPathDescriptives")
if (!exists("treatmentResponse")) {
set.seed(123)
# Step 1: Create the data frame with the first two columns.
treatmentResponse <- data.frame(
patientID = 1:50,
change = round(c(rnorm(25, -40, 20), rnorm(25, 20, 25)))
)
# Step 2: Now that 'treatmentResponse$change' exists, create the responseCategory column.
treatmentResponse$responseCategory <- factor(
ifelse(treatmentResponse$change <= -30, "Partial Response",
ifelse(treatmentResponse$change >= 20, "Progressive Disease", "Stable Disease")
)
)
}
head(treatmentResponse)
#> patientID change responseCategory
#> 1 1 -51 Partial Response
#> 2 2 -45 Partial Response
#> 3 3 -9 Stable Disease
#> 4 4 -39 Partial Response
#> 5 5 -37 Partial Response
#> 6 6 -6 Stable Disease
library(ClinicoPath)
#> Registered S3 method overwritten by 'future':
#> method from
#> all.equal.connection parallelly
#> Warning: replacing previous import 'dplyr::select' by 'jmvcore::select' when
#> loading 'ClinicoPath'
#> Warning: replacing previous import 'cutpointr::roc' by 'pROC::roc' when loading
#> 'ClinicoPath'
#> Warning: replacing previous import 'cutpointr::auc' by 'pROC::auc' when loading
#> 'ClinicoPath'
#> Warning: replacing previous import 'magrittr::extract' by 'tidyr::extract' when
#> loading 'ClinicoPath'
#> Warning in check_dep_version(): ABI version mismatch:
#> lme4 was built with Matrix ABI version 1
#> Current Matrix ABI version is 0
#> Please re-install lme4 from source or restore original 'Matrix' package
#> Warning: replacing previous import 'jmvcore::select' by 'dplyr::select' when
#> loading 'ClinicoPath'
#> Registered S3 methods overwritten by 'ggpp':
#> method from
#> heightDetails.titleGrob ggplot2
#> widthDetails.titleGrob ggplot2
#> Warning: replacing previous import 'DataExplorer::plot_histogram' by
#> 'grafify::plot_histogram' when loading 'ClinicoPath'
#> Warning: replacing previous import 'ROCR::plot' by 'graphics::plot' when
#> loading 'ClinicoPath'
#> Warning: replacing previous import 'dplyr::select' by 'jmvcore::select' when
#> loading 'ClinicoPath'
#> Warning: replacing previous import 'tibble::view' by 'summarytools::view' when
#> loading 'ClinicoPath'
#>
#> Attaching package: 'ClinicoPath'
#> The following object is masked _by_ '.GlobalEnv':
#>
#> treatmentResponse
# data(treatmentResponse, package = "ClinicoPathDescriptives") # if needed
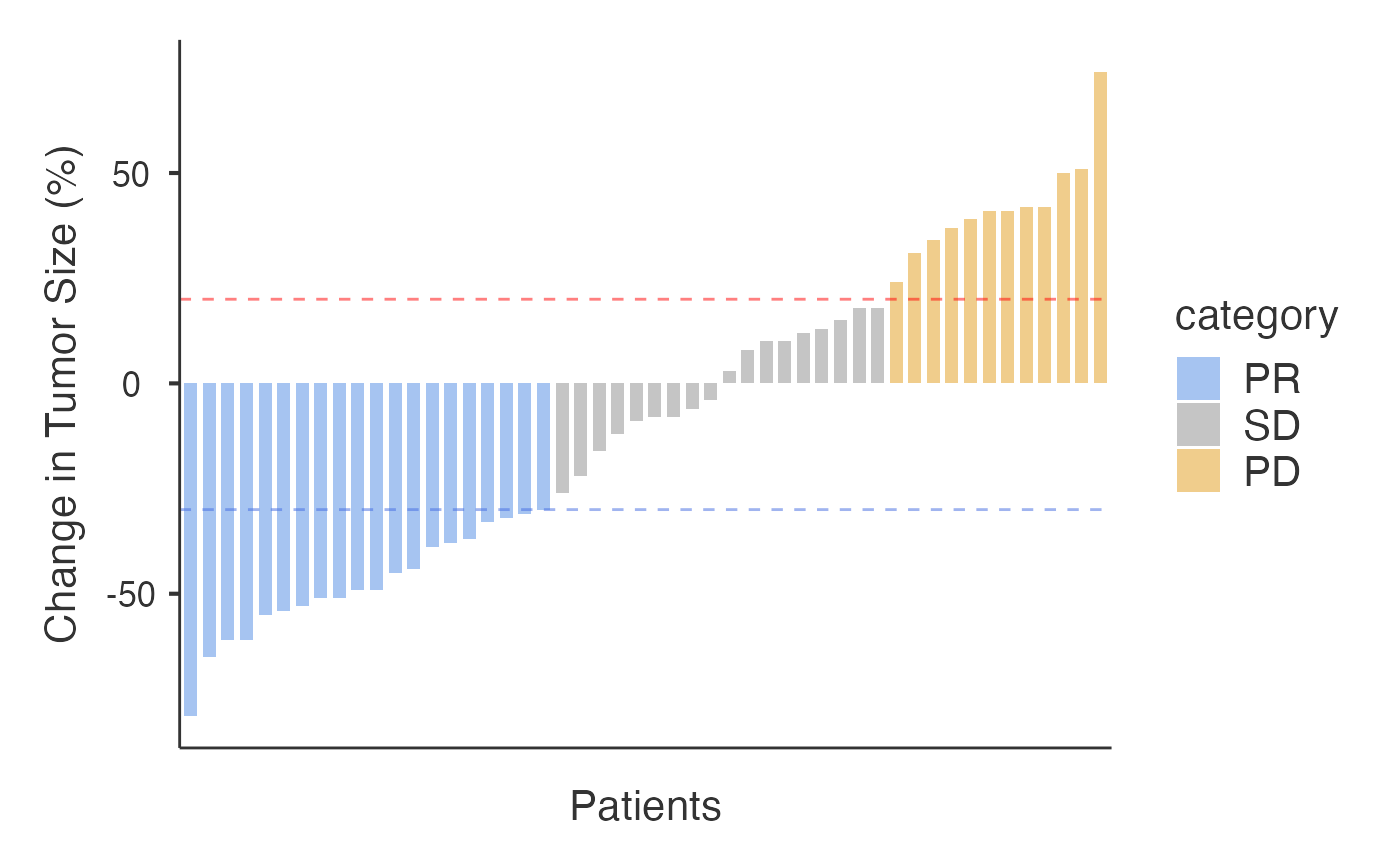
results_waterfall <- ClinicoPath::waterfall(
data = treatmentResponse,
patientID = "patientID",
responseVar = "change",
timeVar = NULL, # Not needed for percentage data
showThresholds = TRUE, # Show +20% and -30% RECIST thresholds
showWaterfallPlot = TRUE
)
# The waterfall function returns a jamovi results object
# The plot is rendered automatically when showWaterfallPlot = TRUE
# Display the results structure
if (!is.null(results_waterfall)) {
print(results_waterfall)
} else {
cat("Results object is NULL - check function execution above\n")
}
#>
#> TREATMENT RESPONSE ANALYSIS
#>
#> Response Categories Based on RECIST v1.1 Criteria
#> ─────────────────────────────────────────────────
#> Category Number of Patients Percentage
#> ─────────────────────────────────────────────────
#> CR ᵃ 0 0%
#> PR ᵇ 20 40%
#> SD ᵈ 18 36%
#> PD ᵉ 12 24%
#> ─────────────────────────────────────────────────
#> ᵃ Complete Response (CR): Complete
#> disappearance of all target lesions.
#> ᵇ Partial Response (PR): At least 30%
#> decrease in sum of target lesions.
#> ᵈ Stable Disease (SD): Neither PR nor PD
#> criteria met.
#> ᵉ Progressive Disease (PD): At least 20%
#> increase in sum of target lesions.
#>
#>
#> Person-Time Analysis
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
#> Response Category Patients % Patients Person-Time % Time Median Time to Response Median Duration
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
#> ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
#>
#>
#> Clinical Response Metrics
#> ────────────────────────────────────────────
#> Metric Value
#> ────────────────────────────────────────────
#> Objective Response Rate (CR+PR) 40%
#> Disease Control Rate (CR+PR+SD) 76%
#> ────────────────────────────────────────────