Continuous Variable Comparisons
Source:vignettes/jjstatsplot-07-continuous-comparisons-legacy.Rmd
jjstatsplot-07-continuous-comparisons-legacy.RmdThis vignette demonstrates functions for analyzing and visualizing continuous variables in the jjstatsplot package. These functions provide comprehensive statistical analyses alongside publication-ready plots.
Between-group comparisons with jjbetweenstats()
The jjbetweenstats() function creates box-violin plots
to compare continuous variables between independent groups. It
automatically performs appropriate statistical tests (parametric,
non-parametric, robust, or Bayesian) and displays the results.
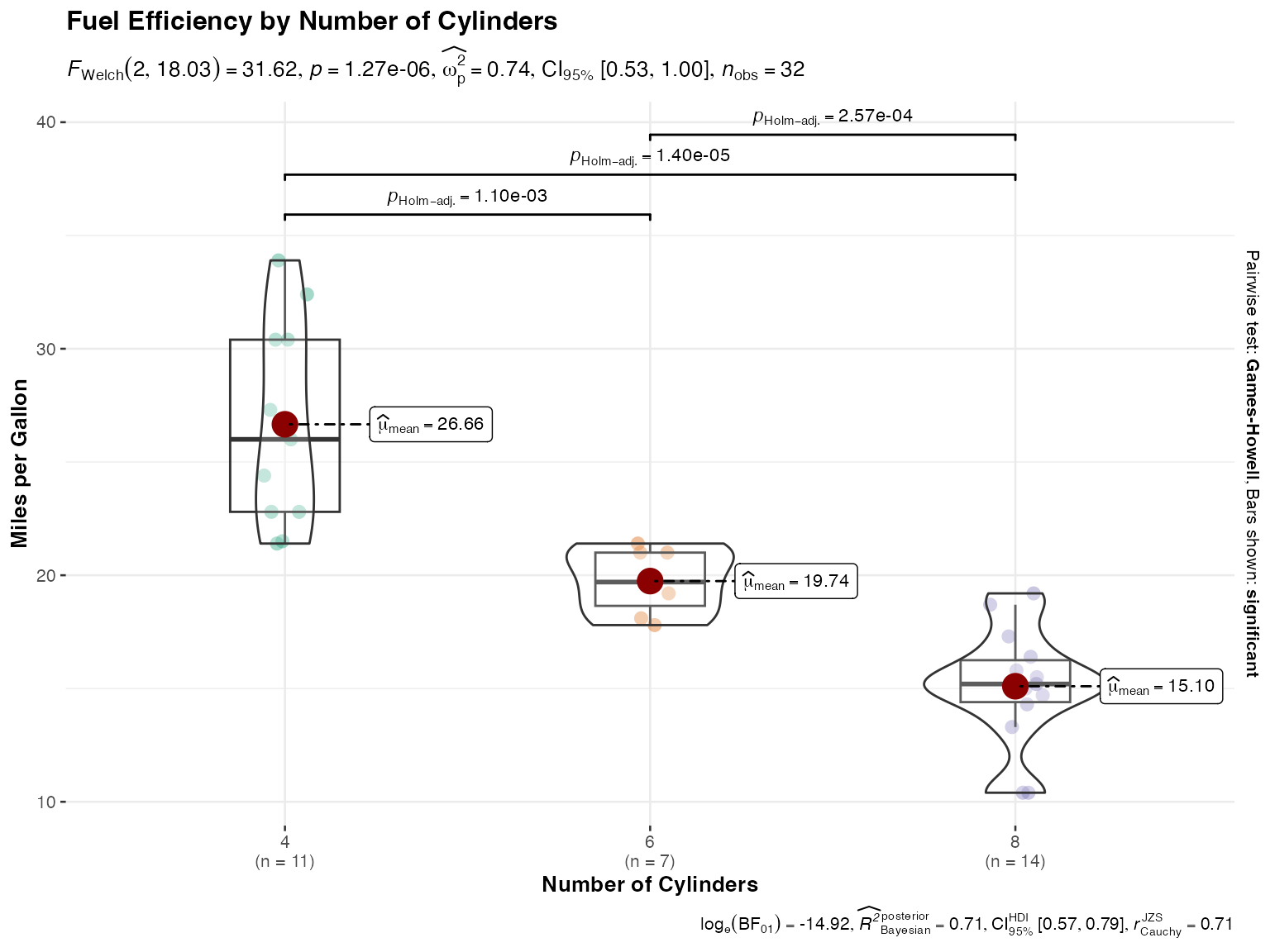
# Underlying function that jjbetweenstats() wraps
ggstatsplot::ggbetweenstats(
data = mtcars,
x = cyl,
y = mpg,
type = "parametric",
pairwise.comparisons = TRUE,
pairwise.display = "significant",
centrality.plotting = TRUE,
title = "Fuel Efficiency by Number of Cylinders",
xlab = "Number of Cylinders",
ylab = "Miles per Gallon"
)
Customizing the analysis type
You can choose different types of statistical analyses:
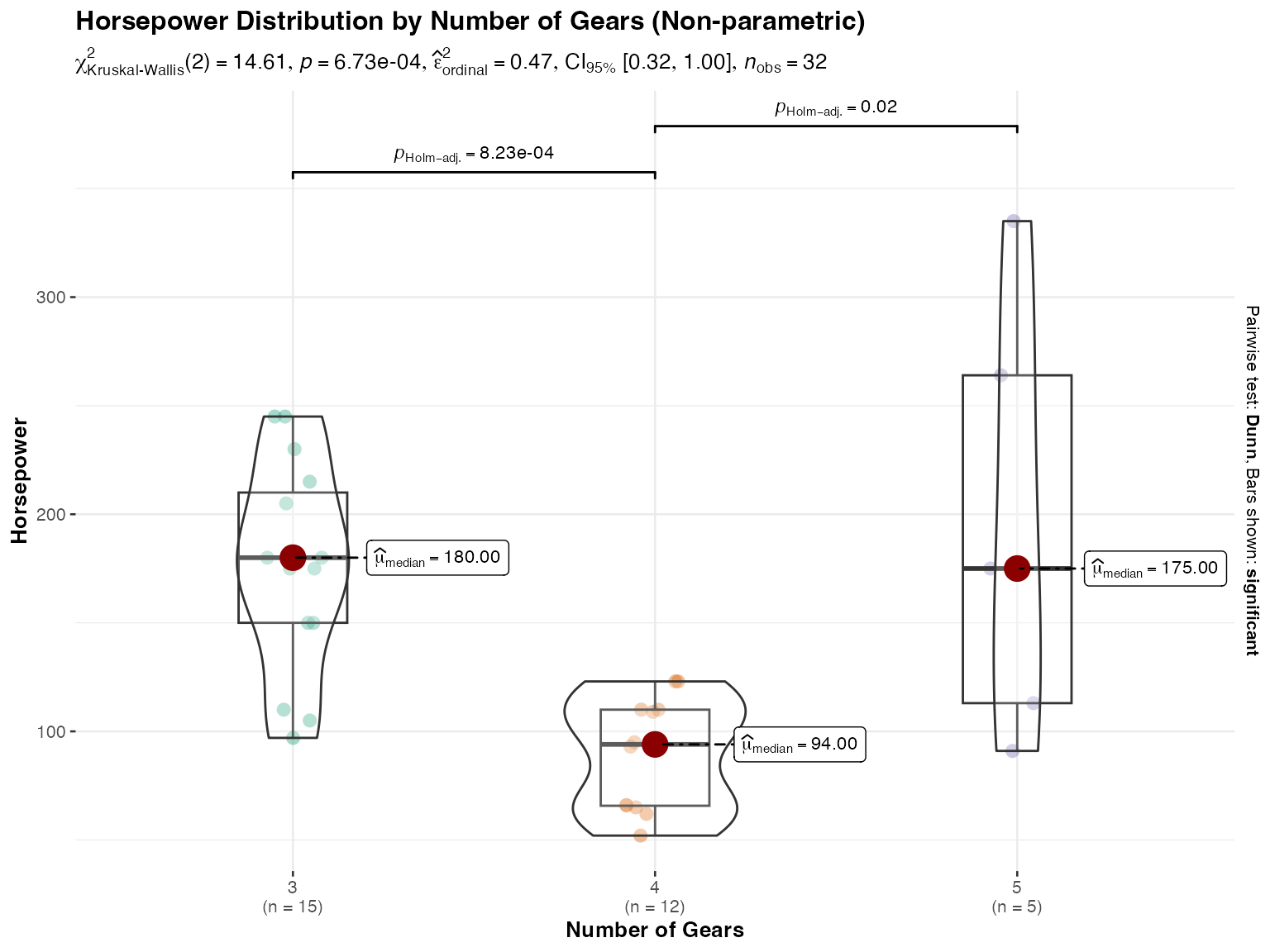
# Non-parametric analysis with Kruskal-Wallis test
ggstatsplot::ggbetweenstats(
data = mtcars,
x = gear,
y = hp,
type = "nonparametric",
centrality.plotting = TRUE,
centrality.type = "nonparametric",
title = "Horsepower Distribution by Number of Gears (Non-parametric)",
xlab = "Number of Gears",
ylab = "Horsepower"
)
Histograms with jjhistostats()
The jjhistostats() function creates histograms with
overlaid density curves and statistical test results against a specified
test value.
# Underlying function that jjhistostats() wraps
ggstatsplot::gghistostats(
data = mtcars,
x = mpg,
type = "parametric",
normal.curve = TRUE,
binwidth = 2,
title = "Distribution of Fuel Efficiency",
xlab = "Miles per Gallon"
)
Grouped histograms
You can also create histograms split by a grouping variable:
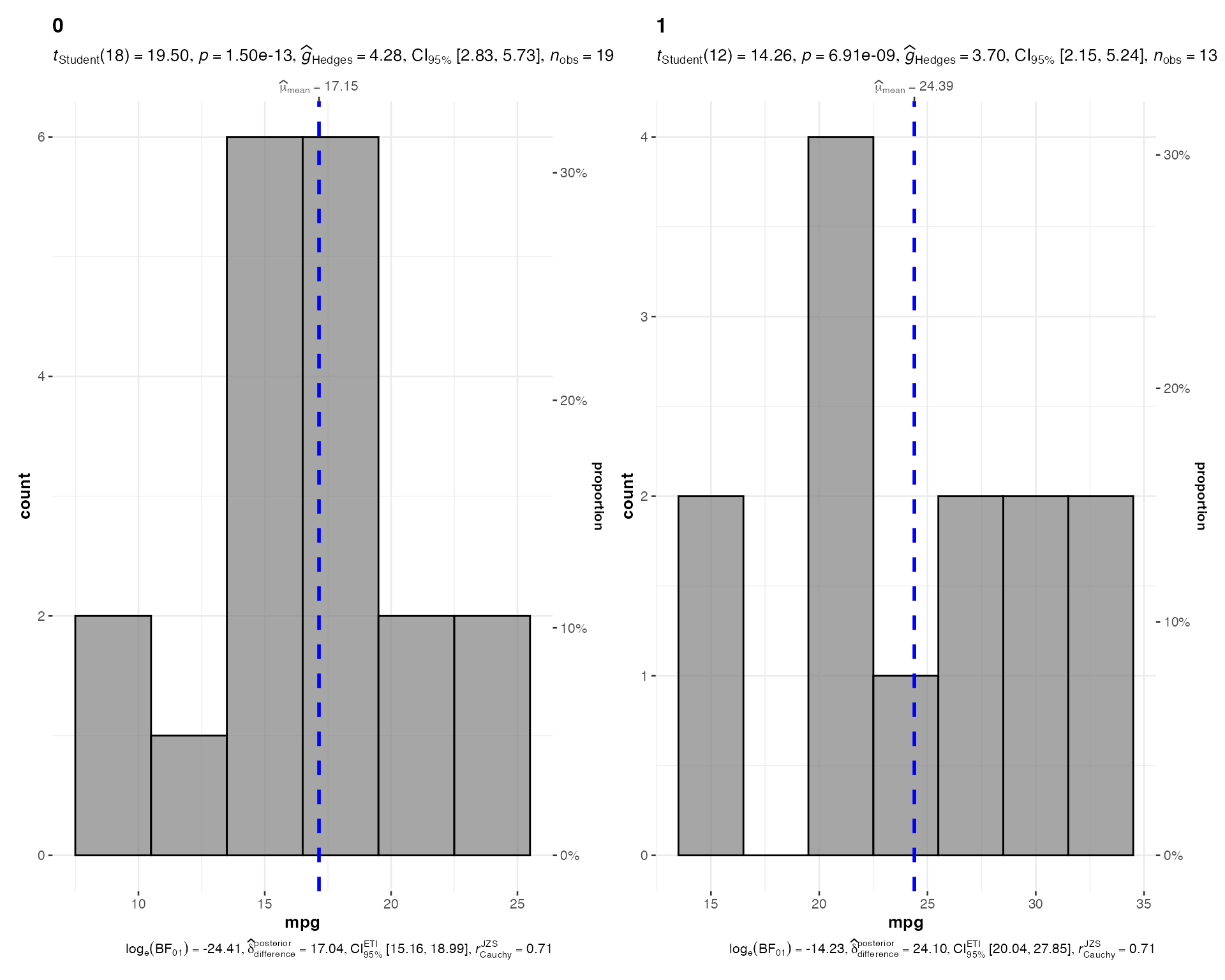
# Grouped histogram
ggstatsplot::grouped_gghistostats(
data = mtcars,
x = mpg,
grouping.var = am,
binwidth = 3,
title.prefix = "Transmission Type",
normal.curve = TRUE
)
Within-group comparisons with jjwithinstats()
The jjwithinstats() function is designed for repeated
measures or paired comparisons. It shows individual trajectories and
performs appropriate paired tests.
# Create paired data for demonstration
library(tidyr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
# Simulate paired measurements (e.g., before and after treatment)
paired_data <- data.frame(
id = rep(1:16, 2),
condition = rep(c("Before", "After"), each = 16),
value = c(mtcars$mpg[1:16], mtcars$mpg[1:16] + rnorm(16, mean = 2, sd = 1))
)
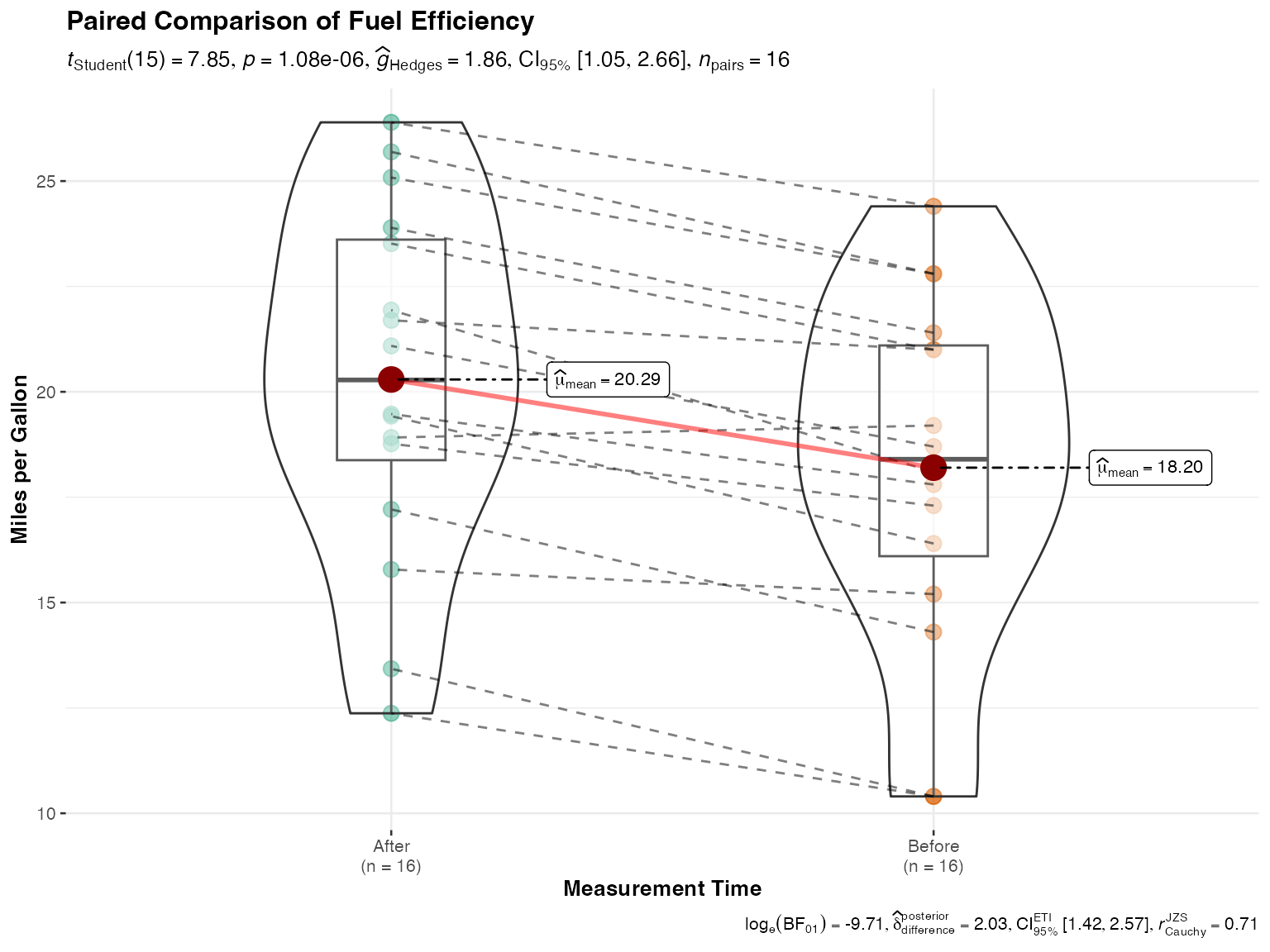
# Underlying function that jjwithinstats() wraps
ggstatsplot::ggwithinstats(
data = paired_data,
x = condition,
y = value,
paired = TRUE,
id = id,
type = "parametric",
pairwise.comparisons = TRUE,
title = "Paired Comparison of Fuel Efficiency",
xlab = "Measurement Time",
ylab = "Miles per Gallon"
)
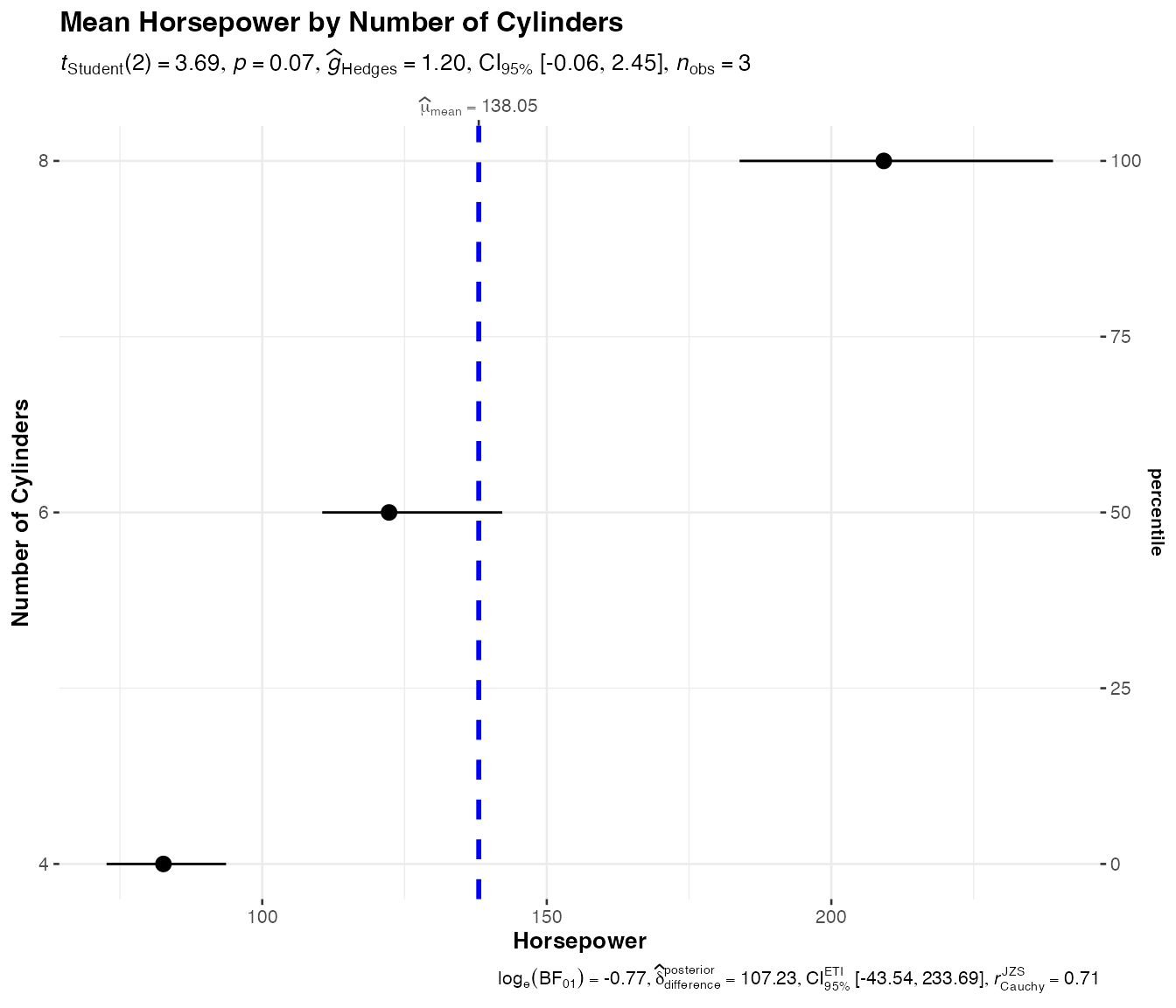
Dot plots with jjdotplotstats()
The jjdotplotstats() function creates Cleveland dot
plots showing group means or medians with confidence intervals. This is
particularly useful for comparing multiple groups when you want to
emphasize the central tendency and uncertainty.
# Underlying function that jjdotplotstats() wraps
ggstatsplot::ggdotplotstats(
data = mtcars,
x = hp,
y = cyl,
type = "parametric",
centrality.plotting = TRUE,
title = "Mean Horsepower by Number of Cylinders",
xlab = "Horsepower",
ylab = "Number of Cylinders"
)
Statistical details
All these functions provide:
-
Automatic statistical testing: Based on the
typeparameter, appropriate tests are selected:- Parametric: t-test, ANOVA, paired t-test
- Non-parametric: Mann-Whitney U, Kruskal-Wallis, Wilcoxon signed-rank
- Robust: Yuen’s trimmed means test, heteroscedastic ANOVA
- Bayesian: Bayes Factor analysis
-
Effect sizes: Appropriate effect size measures are
computed and displayed:
- Cohen’s d, Hedge’s g (for t-tests)
- Eta-squared, omega-squared (for ANOVA)
- r (for non-parametric tests)
- Pairwise comparisons: When comparing multiple groups, post-hoc pairwise comparisons are available with p-value adjustment methods.
Usage in jamovi
These functions are integrated into the jamovi interface where they provide:
- Point-and-click variable selection
- Interactive options for customizing plots
- Automatic handling of missing data
- Export options for plots and results
To use these in jamovi:
- Install the jjstatsplot module from the jamovi library
- Load your dataset
- Navigate to JJStatsPlot → Continuous menu
- Select the appropriate analysis
- Configure options through the user-friendly interface
Advanced customization
While the jamovi interface provides the most common options, R users can access additional customization through the underlying ggstatsplot functions:
# Example with extensive customization
ggstatsplot::ggbetweenstats(
data = mtcars,
x = am,
y = mpg,
type = "parametric",
# Visual customization
violin.args = list(width = 0.5, alpha = 0.2),
boxplot.args = list(width = 0.2, alpha = 0.5),
point.args = list(alpha = 0.5, size = 3, position = position_jitter(width = 0.1)),
# Statistical options
conf.level = 0.99,
effsize.type = "unbiased",
# Theming
ggtheme = ggplot2::theme_minimal(),
# Labels
title = "Fuel Efficiency: Manual vs Automatic Transmission",
subtitle = "99% confidence intervals shown",
xlab = "Transmission Type",
ylab = "Miles per Gallon",
caption = "Data: Motor Trend Car Road Tests"
)
Summary
The continuous comparison functions in jjstatsplot provide:
- Comprehensive statistical analyses with minimal code
- Publication-ready visualizations
- Automatic selection of appropriate statistical tests
- Integration with jamovi’s graphical interface
- Flexibility for advanced R users
These tools make it easy to perform rigorous statistical comparisons while creating informative visualizations suitable for both exploration and presentation.