Creates Raincloud plots to visualize data distributions using ggdist package. Raincloud plots combine three visualization techniques: half-violin plots showing distribution density, box plots showing summary statistics, and dot plots showing individual data points. This provides a comprehensive view of data distribution that reveals patterns traditional box plots might miss, including multimodality and distribution shape. Based on the ggdist R-Bloggers tutorial.
Usage
raincloud(
data,
dep_var,
group_var,
facet_var = NULL,
color_var = NULL,
show_violin = TRUE,
show_boxplot = TRUE,
show_dots = TRUE,
dots_side = "left",
violin_width = 0.7,
box_width = 0.2,
dots_size = 1.2,
alpha_violin = 0.7,
alpha_dots = 0.8,
orientation = "horizontal",
color_palette = "clinical",
plot_theme = "clinical",
plot_title = "Raincloud Plot - Distribution Visualization",
x_label = "",
y_label = "",
show_statistics = FALSE,
show_outliers = FALSE,
outlier_method = "iqr",
normality_test = FALSE,
comparison_test = FALSE,
comparison_method = "auto",
adjust_method = "none",
effect_size = FALSE,
log_transform = FALSE
)Arguments
- data
The data as a data frame.
- dep_var
Continuous variable whose distribution will be visualized in the raincloud plot.
- group_var
Categorical variable for grouping. Each group will have its own raincloud visualization.
- facet_var
Optional variable for creating separate panels. Creates multiple raincloud plots in a grid layout.
- color_var
Optional variable for coloring different elements. If not specified, uses grouping variable.
- show_violin
If TRUE, displays half-violin plot showing probability density distribution.
- show_boxplot
If TRUE, displays box plot with median, quartiles, and outliers.
- show_dots
If TRUE, displays individual data points as dots.
- dots_side
Position of data point dots relative to the violin plot.
- violin_width
Width scaling factor for the violin plot component.
- box_width
Width of the box plot component.
- dots_size
Size of individual data point dots.
- alpha_violin
Transparency level for violin plot (0 = transparent, 1 = opaque).
- alpha_dots
Transparency level for data point dots.
- orientation
Orientation of the plot. Horizontal creates the classic "raincloud" appearance.
- color_palette
Color palette for different groups including GraphPad Prism palettes.
- plot_theme
Overall visual theme for the plot.
- plot_title
Custom title for the raincloud plot.
- x_label
Custom label for X-axis. If empty, uses variable name.
- y_label
Custom label for Y-axis. If empty, uses variable name.
- show_statistics
If TRUE, displays summary statistics table for each group.
- show_outliers
If TRUE, identifies and highlights outliers in the visualization.
- outlier_method
Method for detecting outliers when highlight outliers is enabled.
- normality_test
If TRUE, performs normality tests (Shapiro-Wilk) for each group.
- comparison_test
If TRUE, performs statistical tests to compare groups.
- comparison_method
Statistical test method for comparing groups.
- adjust_method
Adjustment method applied to the reported p-value (useful for multiple comparisons).
- effect_size
If TRUE and two groups are present, reports Cohen's d effect size.
- log_transform
Apply log10 transformation to the Y-axis (requires all positive values).
Value
A results object containing:
results$todo | a html | ||||
results$plot | an image | ||||
results$statistics | a html | ||||
results$outliers | a html | ||||
results$normality | a html | ||||
results$comparison | a html | ||||
results$interpretation | a html |
Examples
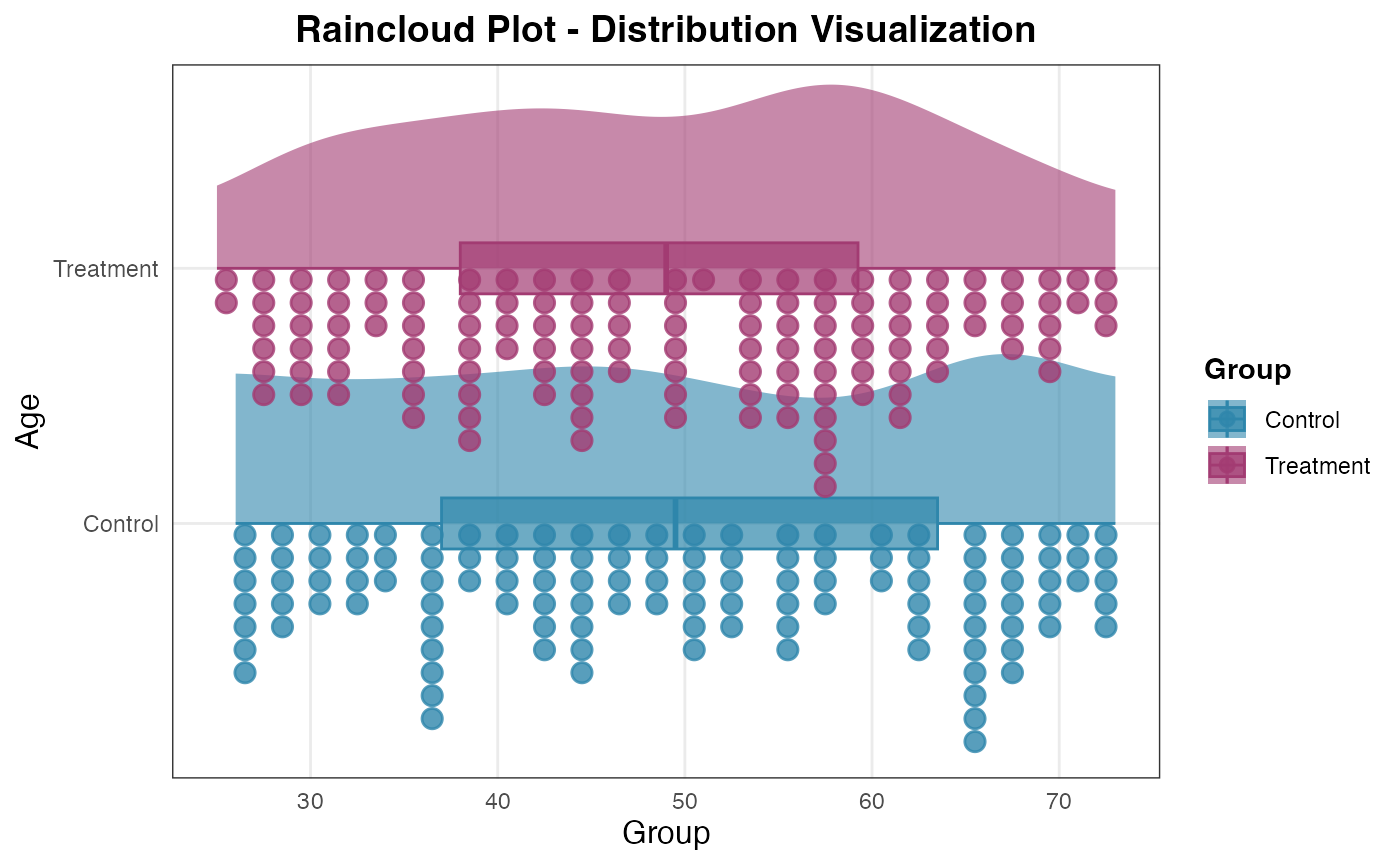
# Load example dataset
data(histopathology)
# Basic raincloud plot
raincloud(
data = histopathology,
dep_var = "Age",
group_var = "Group"
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 248 complete rows (removed 2 with missing values). 2 groups:
#> Control (n=120), Treatment (n=128).
#> Missing by variable:<ul style='margin:6px 0;'>Age: 3 missingGroup: 3
#> missing
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: Age (distribution analysis)Groups: 2 groups defined by
#> GroupObservations: 248 data pointsVisualization: Half-violin (density)
#> + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*
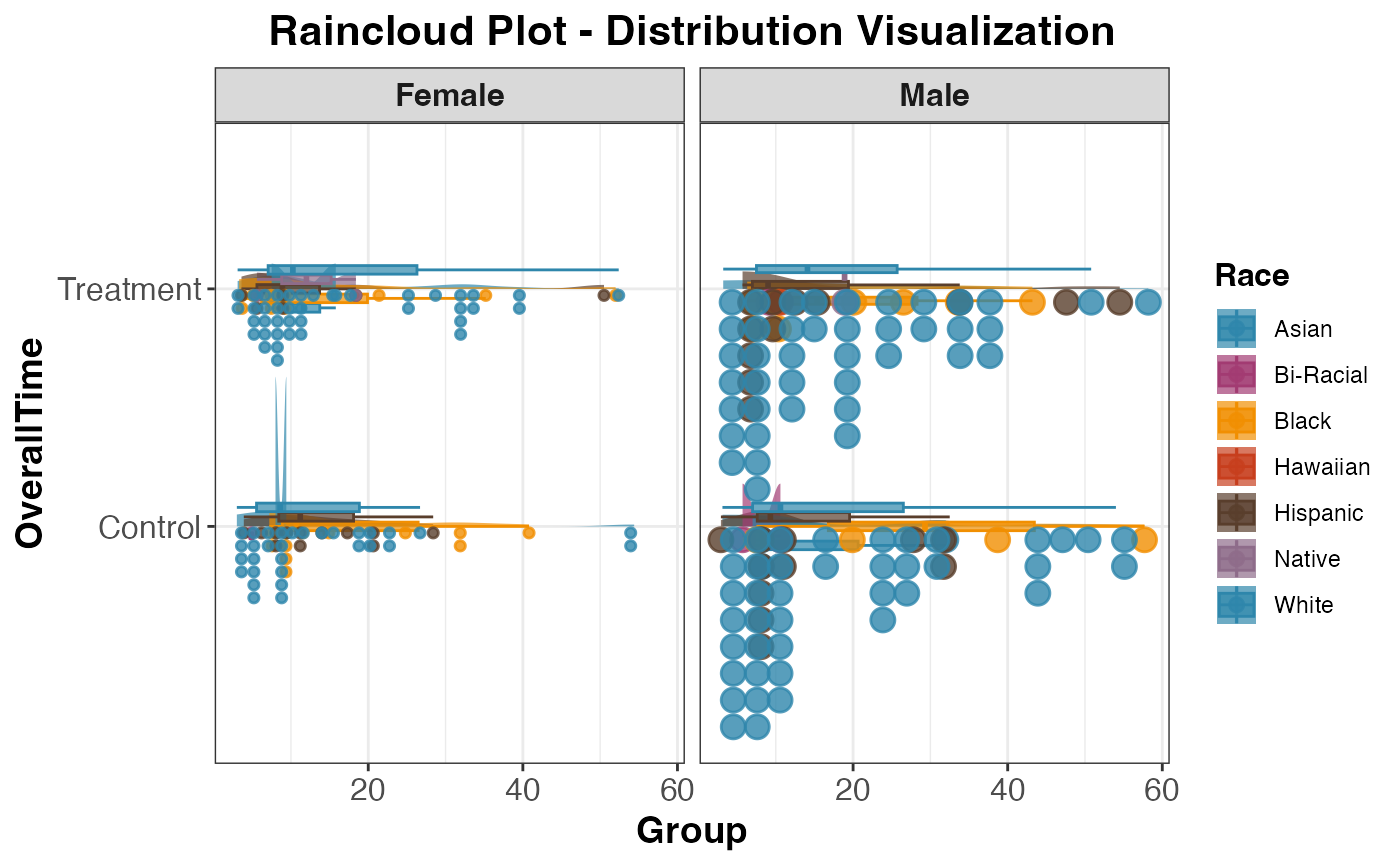
 # Advanced raincloud plot with faceting and custom colors
raincloud(
data = histopathology,
dep_var = "OverallTime",
group_var = "Group",
facet_var = "Sex",
color_var = "Race",
color_palette = "clinical",
plot_theme = "publication"
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 245 complete rows (removed 5 with missing values). 2 groups:
#> Control (n=119), Treatment (n=126).
#> Missing by variable:<ul style='margin:6px 0;'>OverallTime: 7
#> missingGroup: 6 missingSex: 6 missingRace: 6 missing
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: OverallTime (distribution analysis)Groups: 2 groups defined
#> by GroupObservations: 245 data pointsVisualization: Half-violin
#> (density) + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*
# Advanced raincloud plot with faceting and custom colors
raincloud(
data = histopathology,
dep_var = "OverallTime",
group_var = "Group",
facet_var = "Sex",
color_var = "Race",
color_palette = "clinical",
plot_theme = "publication"
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 245 complete rows (removed 5 with missing values). 2 groups:
#> Control (n=119), Treatment (n=126).
#> Missing by variable:<ul style='margin:6px 0;'>OverallTime: 7
#> missingGroup: 6 missingSex: 6 missingRace: 6 missing
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: OverallTime (distribution analysis)Groups: 2 groups defined
#> by GroupObservations: 245 data pointsVisualization: Half-violin
#> (density) + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*
 # Statistical analysis with outlier detection
raincloud(
data = histopathology,
dep_var = "Age",
group_var = "Group",
show_statistics = TRUE,
show_outliers = TRUE,
outlier_method = "iqr",
normality_test = TRUE,
comparison_test = TRUE
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 248 complete rows (removed 2 with missing values). 2 groups:
#> Control (n=120), Treatment (n=128).
#> Missing by variable:<ul style='margin:6px 0;'>Age: 3 missingGroup: 3
#> missing
#>
#> <div style='background-color: #f8f9fa; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #495057; margin-top:
#> 0;'>📊 Distribution Summary Statistics
#>
#> <table style='width: 100%; border-collapse: collapse; font-family:
#> Arial, sans-serif;'><tr style='background-color: #6c757d; color:
#> white;'><th style='padding: 8px; border: 1px solid #dee2e6;'>Group<th
#> style='padding: 8px; border: 1px solid #dee2e6;'>N<th style='padding:
#> 8px; border: 1px solid #dee2e6;'>Mean<th style='padding: 8px; border:
#> 1px solid #dee2e6;'>Median<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>SD<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>IQR<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>Range<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>Skewness<tr style='background-color:
#> #f8f9fa;'>Control12049.82549.514.41526.526 - 73-0.017<tr
#> style='background-color: #ffffff;'>Treatment12848.9694913.25621.2525 -
#> 73-0.046<p style='font-size: 12px; color: #6c757d; margin-top:
#> 15px;'>*IQR = Interquartile Range, SD = Standard Deviation. Skewness:
#> 0 = symmetric, >0 = right-skewed, <0 = left-skewed.*
#>
#> <div style='background-color: #fff3cd; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #856404; margin-top:
#> 0;'>⚠️ Outlier Detection (Iqr Method)
#>
#> Control: 0 outliers detectedTreatment: 0 outliers detected
#>
#> Total outliers across all groups: 0
#>
#> <p style='font-size: 12px; color: #856404; margin-top: 15px;'>*IQR
#> Method: Values beyond 1.5 × IQR from Q1/Q3. Consider investigating
#> these points for data quality or interesting patterns.*
#>
#> <div style='background-color: #d1ecf1; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #0c5460; margin-top:
#> 0;'>📈 Normality Tests (Shapiro-Wilk)
#>
#> <table style='width: 100%; border-collapse: collapse;'><tr
#> style='background-color: #e0e0e0;'><th style='padding: 8px; border:
#> 1px solid #ddd;'>Group<th style='padding: 8px; border: 1px solid
#> #ddd;'>W Statistic<th style='padding: 8px; border: 1px solid
#> #ddd;'>P-value<th style='padding: 8px; border: 1px solid
#> #ddd;'>InterpretationControl0.94321e-04Non-normalTreatment0.95947e-04Non-normal<p
#> style='font-size: 12px; color: #0c5460; margin-top:
#> 15px;'>*Shapiro-Wilk test: p > 0.05 suggests normal distribution.
#> Valid for sample sizes 3-5000.*
#>
#> <div style='background-color: #f3e5f5; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #7b1fa2; margin-top:
#> 0;'>📊 Group Comparison Test
#>
#> <table style='width: 100%; border-collapse: collapse;'><td
#> style='padding: 8px; border: 1px solid #ddd;'>Test Method:<td
#> style='padding: 8px; border: 1px solid #ddd;'>Wilcoxon<td
#> style='padding: 8px; border: 1px solid #ddd;'>Test Statistic:<td
#> style='padding: 8px; border: 1px solid #ddd;'>W = 7955.5<td
#> style='padding: 8px; border: 1px solid #ddd;'>P-value:<td
#> style='padding: 8px; border: 1px solid #ddd;'>0.6261<td
#> style='padding: 8px; border: 1px solid #ddd;'>Result:<td
#> style='padding: 8px; border: 1px solid #ddd;'>Not significant<p
#> style='font-size: 12px; color: #7b1fa2; margin-top: 15px;'>** p <
#> 0.05, ** p < 0.01, *** p < 0.001. Automatically selected {test_method}
#> based on data characteristics.*
#>
#> <div style='background-color: #e3f2fd; padding: 15px; border-radius:
#> 5px; margin: 10px 0;'><h4 style='margin-top: 0; color: #1976d2;'>📝
#> Copy-Ready Result
#>
#> <p style='font-family: monospace; background: white; padding: 10px;
#> border: 1px solid #ddd; border-radius: 3px;'>The Wilcoxon test
#> comparing Control vs Treatment showed not significant differences (p =
#> 0.626). No significant differences detected between groups.
#>
#> <small style='color: #666;'>💡 This sentence is ready to copy into
#> your research report
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: Age (distribution analysis)Groups: 2 groups defined by
#> GroupObservations: 248 data pointsVisualization: Half-violin (density)
#> + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*
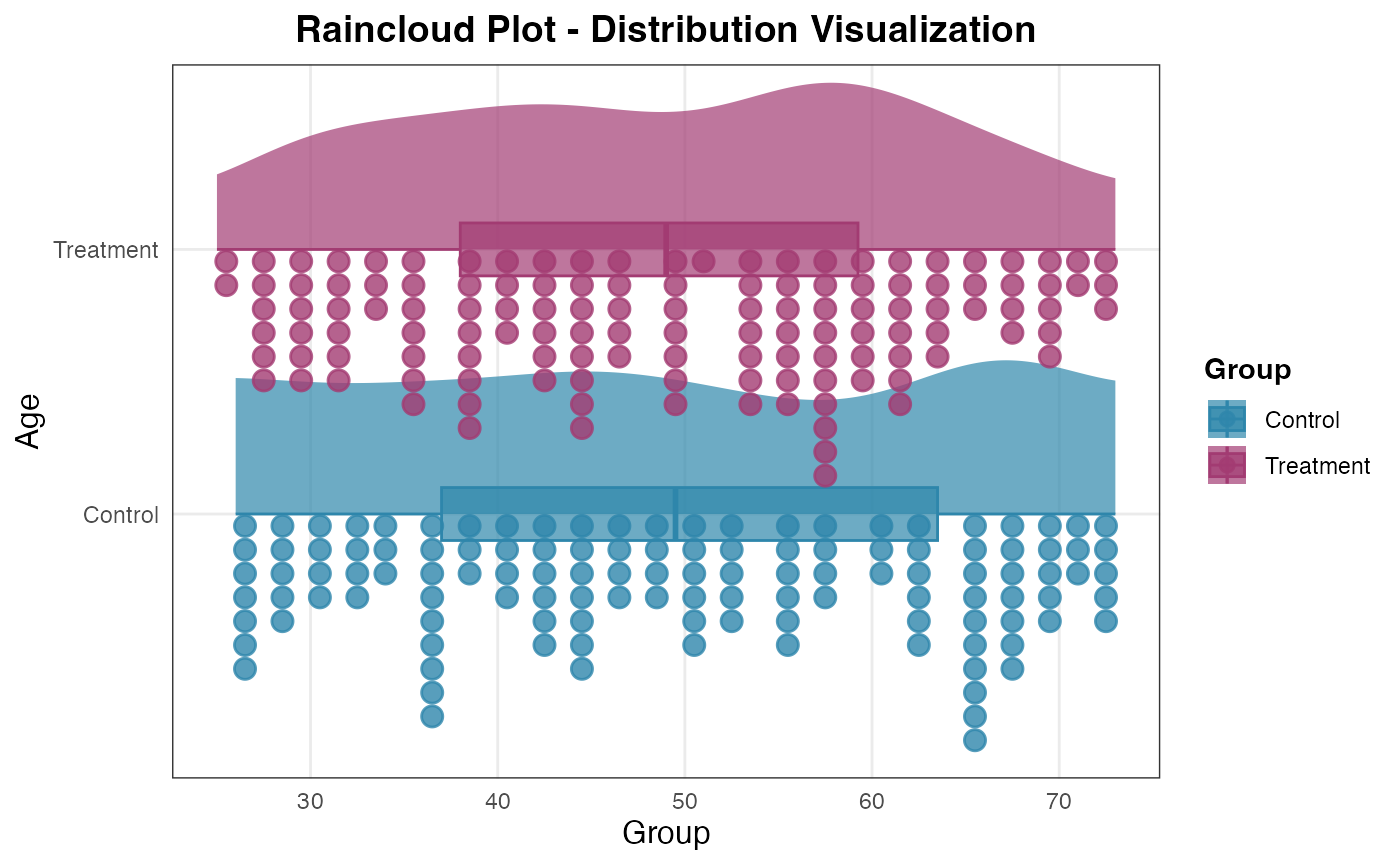
# Statistical analysis with outlier detection
raincloud(
data = histopathology,
dep_var = "Age",
group_var = "Group",
show_statistics = TRUE,
show_outliers = TRUE,
outlier_method = "iqr",
normality_test = TRUE,
comparison_test = TRUE
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 248 complete rows (removed 2 with missing values). 2 groups:
#> Control (n=120), Treatment (n=128).
#> Missing by variable:<ul style='margin:6px 0;'>Age: 3 missingGroup: 3
#> missing
#>
#> <div style='background-color: #f8f9fa; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #495057; margin-top:
#> 0;'>📊 Distribution Summary Statistics
#>
#> <table style='width: 100%; border-collapse: collapse; font-family:
#> Arial, sans-serif;'><tr style='background-color: #6c757d; color:
#> white;'><th style='padding: 8px; border: 1px solid #dee2e6;'>Group<th
#> style='padding: 8px; border: 1px solid #dee2e6;'>N<th style='padding:
#> 8px; border: 1px solid #dee2e6;'>Mean<th style='padding: 8px; border:
#> 1px solid #dee2e6;'>Median<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>SD<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>IQR<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>Range<th style='padding: 8px; border: 1px solid
#> #dee2e6;'>Skewness<tr style='background-color:
#> #f8f9fa;'>Control12049.82549.514.41526.526 - 73-0.017<tr
#> style='background-color: #ffffff;'>Treatment12848.9694913.25621.2525 -
#> 73-0.046<p style='font-size: 12px; color: #6c757d; margin-top:
#> 15px;'>*IQR = Interquartile Range, SD = Standard Deviation. Skewness:
#> 0 = symmetric, >0 = right-skewed, <0 = left-skewed.*
#>
#> <div style='background-color: #fff3cd; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #856404; margin-top:
#> 0;'>⚠️ Outlier Detection (Iqr Method)
#>
#> Control: 0 outliers detectedTreatment: 0 outliers detected
#>
#> Total outliers across all groups: 0
#>
#> <p style='font-size: 12px; color: #856404; margin-top: 15px;'>*IQR
#> Method: Values beyond 1.5 × IQR from Q1/Q3. Consider investigating
#> these points for data quality or interesting patterns.*
#>
#> <div style='background-color: #d1ecf1; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #0c5460; margin-top:
#> 0;'>📈 Normality Tests (Shapiro-Wilk)
#>
#> <table style='width: 100%; border-collapse: collapse;'><tr
#> style='background-color: #e0e0e0;'><th style='padding: 8px; border:
#> 1px solid #ddd;'>Group<th style='padding: 8px; border: 1px solid
#> #ddd;'>W Statistic<th style='padding: 8px; border: 1px solid
#> #ddd;'>P-value<th style='padding: 8px; border: 1px solid
#> #ddd;'>InterpretationControl0.94321e-04Non-normalTreatment0.95947e-04Non-normal<p
#> style='font-size: 12px; color: #0c5460; margin-top:
#> 15px;'>*Shapiro-Wilk test: p > 0.05 suggests normal distribution.
#> Valid for sample sizes 3-5000.*
#>
#> <div style='background-color: #f3e5f5; padding: 20px; border-radius:
#> 8px; margin-bottom: 20px;'><h3 style='color: #7b1fa2; margin-top:
#> 0;'>📊 Group Comparison Test
#>
#> <table style='width: 100%; border-collapse: collapse;'><td
#> style='padding: 8px; border: 1px solid #ddd;'>Test Method:<td
#> style='padding: 8px; border: 1px solid #ddd;'>Wilcoxon<td
#> style='padding: 8px; border: 1px solid #ddd;'>Test Statistic:<td
#> style='padding: 8px; border: 1px solid #ddd;'>W = 7955.5<td
#> style='padding: 8px; border: 1px solid #ddd;'>P-value:<td
#> style='padding: 8px; border: 1px solid #ddd;'>0.6261<td
#> style='padding: 8px; border: 1px solid #ddd;'>Result:<td
#> style='padding: 8px; border: 1px solid #ddd;'>Not significant<p
#> style='font-size: 12px; color: #7b1fa2; margin-top: 15px;'>** p <
#> 0.05, ** p < 0.01, *** p < 0.001. Automatically selected {test_method}
#> based on data characteristics.*
#>
#> <div style='background-color: #e3f2fd; padding: 15px; border-radius:
#> 5px; margin: 10px 0;'><h4 style='margin-top: 0; color: #1976d2;'>📝
#> Copy-Ready Result
#>
#> <p style='font-family: monospace; background: white; padding: 10px;
#> border: 1px solid #ddd; border-radius: 3px;'>The Wilcoxon test
#> comparing Control vs Treatment showed not significant differences (p =
#> 0.626). No significant differences detected between groups.
#>
#> <small style='color: #666;'>💡 This sentence is ready to copy into
#> your research report
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: Age (distribution analysis)Groups: 2 groups defined by
#> GroupObservations: 248 data pointsVisualization: Half-violin (density)
#> + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*
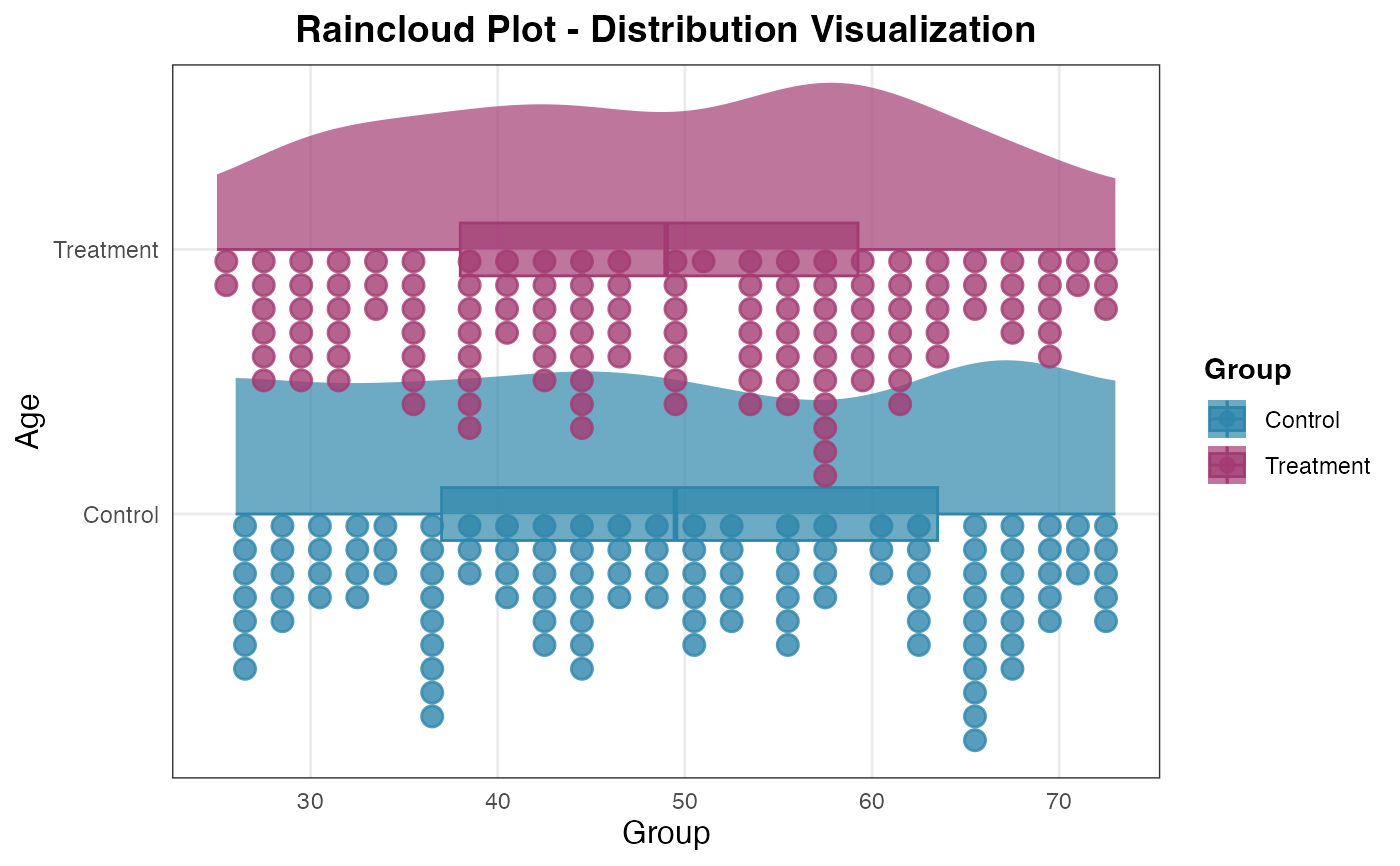 # Customized visualization components
raincloud(
data = histopathology,
dep_var = "Age",
group_var = "Group",
show_violin = TRUE,
show_boxplot = TRUE,
show_dots = TRUE,
dots_side = "left",
orientation = "horizontal",
violin_width = 0.8,
alpha_violin = 0.6
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 248 complete rows (removed 2 with missing values). 2 groups:
#> Control (n=120), Treatment (n=128).
#> Missing by variable:<ul style='margin:6px 0;'>Age: 3 missingGroup: 3
#> missing
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: Age (distribution analysis)Groups: 2 groups defined by
#> GroupObservations: 248 data pointsVisualization: Half-violin (density)
#> + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*
# Customized visualization components
raincloud(
data = histopathology,
dep_var = "Age",
group_var = "Group",
show_violin = TRUE,
show_boxplot = TRUE,
show_dots = TRUE,
dots_side = "left",
orientation = "horizontal",
violin_width = 0.8,
alpha_violin = 0.6
)
#>
#> RAINCLOUD PLOT
#>
#> <div
#> style='background-color:#f8f9fa;padding:12px;border-radius:8px;margin-bottom:12px;'>Data
#> summary: 248 complete rows (removed 2 with missing values). 2 groups:
#> Control (n=120), Treatment (n=128).
#> Missing by variable:<ul style='margin:6px 0;'>Age: 3 missingGroup: 3
#> missing
#>
#> <div style='background-color: #e8f5e8; padding: 20px; border-radius:
#> 8px;'><h3 style='color: #2e7d32; margin-top: 0;'>📋 Raincloud Plot
#> Interpretation Guide
#>
#> <h4 style='color: #2e7d32;'>Plot Summary:
#>
#> Variable: Age (distribution analysis)Groups: 2 groups defined by
#> GroupObservations: 248 data pointsVisualization: Half-violin (density)
#> + Box plot (quartiles) + Data points (individual values)<h4
#> style='color: #2e7d32;'>How to Read Raincloud Plots:
#>
#> Half-Violin: Shows probability density - wider areas indicate more
#> data pointsBox Plot: Shows median (line), quartiles (box), and
#> outliers (points)Data Points: Individual observations reveal
#> fine-grained patternsShape Patterns: Symmetric, skewed, bimodal, or
#> multimodal distributions<h4 style='color: #2e7d32;'>Distribution
#> Patterns to Look For:
#>
#> Symmetry: Bell-shaped density indicates normal distributionSkewness:
#> Tail extending to one side (left-skewed or right-skewed)Multimodality:
#> Multiple peaks suggest subgroups within dataOutliers: Points far from
#> the main distributionSpread: Width of distribution indicates
#> variability<h4 style='color: #2e7d32;'>Clinical/Research Applications:
#>
#> Biomarker Analysis: Compare distributions across patient
#> groupsTreatment Effects: Visualize before/after treatment
#> distributionsQuality Control: Identify unusual patterns in laboratory
#> valuesSubgroup Discovery: Detect hidden subpopulations in data<p
#> style='font-size: 12px; color: #2e7d32; margin-top: 15px;'>*💡
#> Raincloud plots reveal distribution nuances that traditional box plots
#> miss, making them ideal for exploratory data analysis and
#> publication-quality visualizations.*